

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140022.4 - phase: 0
(205 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
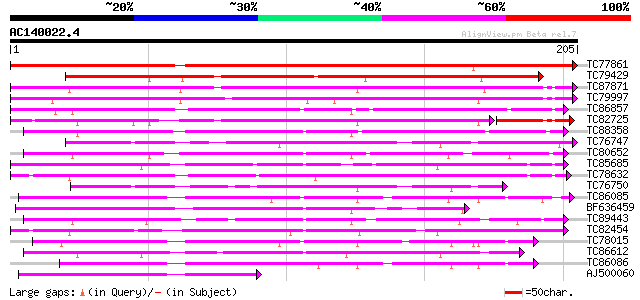
Score E
Sequences producing significant alignments: (bits) Value
TC77861 weakly similar to GP|13899081|gb|AAK48962.1 Unknown prot... 342 6e-95
TC79429 similar to PIR|T03803|T03803 tumor-related protein clon... 155 8e-39
TC87871 similar to PIR|T03803|T03803 tumor-related protein clon... 130 3e-31
TC79997 similar to PIR|T03803|T03803 tumor-related protein clon... 125 9e-30
TC86857 similar to GP|14161088|emb|CAB76907. alpha-fucosidase {C... 100 7e-22
TC82725 weakly similar to PIR|T07871|T07871 miraculin homolog r... 87 7e-19
TC88358 proteinase inhibitor 20 [Medicago truncatula] 89 9e-19
TC76747 similar to GP|20269067|emb|CAD29731. protease inhibitor ... 88 3e-18
TC80652 similar to GP|14161088|emb|CAB76907. alpha-fucosidase {C... 87 4e-18
TC85685 similar to GP|14161088|emb|CAB76907. alpha-fucosidase {C... 81 2e-16
TC78632 similar to GP|9367042|gb|AAF87095.1| trypsin inhibitor {... 78 2e-15
TC76750 similar to GP|20269067|emb|CAD29731. protease inhibitor ... 76 8e-15
TC86085 74 5e-14
BF636459 73 9e-14
TC89443 similar to GP|7208767|emb|CAB76906.1 alpha-fucosidase {C... 65 2e-11
TC82454 weakly similar to GP|3551243|dbj|BAA32820.1 Water-Solubl... 64 3e-11
TC78015 similar to GP|12721693|gb|AAK03411.1 RbsC {Pasteurella m... 63 7e-11
TC86612 weakly similar to GP|7208767|emb|CAB76906.1 alpha-fucosi... 62 2e-10
TC86086 similar to GP|15155442|gb|AAK86324.1 AGR_C_899p {Agrobac... 60 8e-10
AJ500060 50 8e-07
>TC77861 weakly similar to GP|13899081|gb|AAK48962.1 Unknown protein
{Arabidopsis thaliana}, partial (39%)
Length = 871
Score = 342 bits (877), Expect = 6e-95
Identities = 169/206 (82%), Positives = 182/206 (88%), Gaps = 1/206 (0%)
Frame = +3
Query: 1 MKTSFLAFSIIFLAFICKTFAAPEPVLDISGKKVTTGVKYYILPVIRGKGGGLTVVNENN 60
MKTSFLAFSI+ LAFICKT AAPEPVLDISGK++TTGVKYYILPVIRGKGGGLTV N
Sbjct: 51 MKTSFLAFSILCLAFICKTIAAPEPVLDISGKQLTTGVKYYILPVIRGKGGGLTVANHGE 230
Query: 61 LNGNNNTCPLYVLQEKLEVKNGQAVTFTPYNAKKGVILTSTDLNIKSYVTKTTCAQSQVW 120
NN TCPLYV+QEKLEVKNG+AVTFTPYNAK+GVILTSTDLNIKS+VTKT C Q+QVW
Sbjct: 231 ---NNQTCPLYVVQEKLEVKNGEAVTFTPYNAKQGVILTSTDLNIKSFVTKTKCPQTQVW 401
Query: 121 KLNKVLSGVWFLATGGVEGNPGFDTIFNWFKIEKADKDYVFSFCPS-VCKCQTLCRELGL 179
KL K L+GVWFLATGGVEGNP T+ NWFKIEKADKDYV SFCP+ CKCQTLCRELGL
Sbjct: 402 KLLKELTGVWFLATGGVEGNPSMATVGNWFKIEKADKDYVLSFCPAEACKCQTLCRELGL 581
Query: 180 YVYDHGKKHLALSDQVPSFRVVFKRA 205
+V D G KHLALSDQ+PSFRVVFKRA
Sbjct: 582 FVDDKGNKHLALSDQIPSFRVVFKRA 659
>TC79429 similar to PIR|T03803|T03803 tumor-related protein clone NF34 -
common tobacco, partial (42%)
Length = 807
Score = 155 bits (393), Expect = 8e-39
Identities = 89/181 (49%), Positives = 113/181 (62%), Gaps = 8/181 (4%)
Frame = +2
Query: 21 AAPEPVLDISGKKVTTGVKYYILPVIRGK--GGGLTVVNENNL---NGNNNTCPLYVLQE 75
A+PE V+D GKKV GV YYI PV G G VV + N TCPL V+
Sbjct: 95 ASPEQVVDTEGKKVRAGVDYYIRPVPTTPCDGRGPCVVGSGFVLIARSPNETCPLNVVV- 271
Query: 76 KLEVKNGQAVTFTPYNAKKGVILTSTDLNIKSYVTKTTCAQSQVWKLNKVLS--GVWFLA 133
+E GQ VTFTP N KKGVI STDLNIK+ + T+C +S +W L+ S G WF+
Sbjct: 272 -VEGFRGQGVTFTPVNPKKGVIRVSTDLNIKTSLN-TSCEESTIWTLDDFDSSTGQWFVT 445
Query: 134 TGGVEGNPGFDTIFNWFKIEKADKDYVFSFCPSVCK-CQTLCRELGLYVYDHGKKHLALS 192
TGGV GNPG DT+ NWFKIEK + DY F FCP+VC C+ +CR +G++ +G + +AL+
Sbjct: 446 TGGVLGNPGKDTVDNWFKIEKYEDDYKFVFCPTVCNFCKVMCRNVGIFRDSNGNQRVALT 625
Query: 193 D 193
D
Sbjct: 626 D 628
>TC87871 similar to PIR|T03803|T03803 tumor-related protein clone NF34 -
common tobacco, partial (36%)
Length = 905
Score = 130 bits (328), Expect = 3e-31
Identities = 88/217 (40%), Positives = 118/217 (53%), Gaps = 12/217 (5%)
Frame = +3
Query: 1 MKTSFLAFSIIFLAFICKTF-----AAPEPVLDISGKKVTTGVKYYILPVIRGKGGGLTV 55
MK + LAF +F + A+ E V+D GKK+ YYI+PV K G
Sbjct: 15 MKNTLLAFFFLFTFLSSQPLLGAAEASNEQVVDTLGKKLRADANYYIIPVPIYKCGPYGK 194
Query: 56 VNENN----LNGNNNTCPLYVLQEKLEVKNGQAVTFTPYNAKKGVILTSTDLNIKSYVTK 111
+ L TCPL V+ ++ +TF P N KKGVI STDLNIK
Sbjct: 195 CRSSGSSLALASIGKTCPLDVVV--VDRYQALPLTFIPVNPKKGVIRVSTDLNIKFSSRA 368
Query: 112 TTCAQSQVWKLNK--VLSGVWFLATGGVEGNPGFDTIFNWFKIEKADKDYVFSFCPSVCK 169
T S VWKL++ V WF+ GGV GNPG++TI NWFKIEK Y FCPSV +
Sbjct: 369 TCLHHSMVWKLDRFNVSKRQWFITIGGVAGNPGWETINNWFKIEKYGDAYKLVFCPSVVQ 548
Query: 170 C-QTLCRELGLYVYDHGKKHLALSDQVPSFRVVFKRA 205
+ +C+++G++V ++G K LALSD VP +V F++A
Sbjct: 549 SFKHMCKDVGVFVDENGNKRLALSD-VP-LKVKFQQA 653
>TC79997 similar to PIR|T03803|T03803 tumor-related protein clone NF34 -
common tobacco, partial (48%)
Length = 801
Score = 125 bits (315), Expect = 9e-30
Identities = 82/216 (37%), Positives = 119/216 (54%), Gaps = 11/216 (5%)
Frame = +2
Query: 1 MKTSFLAFSIIFLA----FICKTFAAPEPVLDISGKKVTTGVKYYILPVIRGKGGGLTVV 56
MK + LAF ++F + A+P+ V+D SGKK+ YYI+P G
Sbjct: 41 MKGTMLAFLLLFALSSQPLLGSAEASPDQVIDTSGKKLRADTNYYIIPAKPFTTCGFVSC 220
Query: 57 NENN---LNGNNNTCPLYVLQEKLEVKNGQAVTFTPYNAKKGVILTSTDLNIK--SYVTK 111
+ L +CPL V+ K G + FTP N KKGV+ STDLNIK +
Sbjct: 221 FNSGGIALETVGESCPLDVVVVKHN--QGLPLRFTPVNNKKGVVRVSTDLNIKFSNDAYD 394
Query: 112 TTCAQ-SQVWKLNKVLSGVWFLATGGVEGNPGFDTIFNWFKIEKADKDYVFSFCPSVC-K 169
+ C S VWK++ F+ T GV GNPG +TI NWF+IEK + Y +CP+VC
Sbjct: 395 SRCPNHSLVWKIDPFSKEETFVTTNGVLGNPGSNTIHNWFQIEKYEDAYKLVYCPNVCPS 574
Query: 170 CQTLCRELGLYVYDHGKKHLALSDQVPSFRVVFKRA 205
C+ +C+++G+YVY + + LAL++ VP F+V F++A
Sbjct: 575 CKHVCKDIGIYVYKYREMRLALTN-VP-FKVKFQKA 676
>TC86857 similar to GP|14161088|emb|CAB76907. alpha-fucosidase {Cicer
arietinum}, partial (41%)
Length = 860
Score = 99.8 bits (247), Expect = 7e-22
Identities = 77/210 (36%), Positives = 106/210 (49%), Gaps = 8/210 (3%)
Frame = +1
Query: 1 MKTSFLAFSIIFLAF-ICKTFA---APEPVLDISGKKVTTGVKYYILPVIRGKGGGLTVV 56
MK +FL S+I A IC + A E V D +G + G +YI+P I G GG +
Sbjct: 79 MKPTFLTLSLILFALTICFSLAFAQVSEQVFDTNGNPIFPGGTFYIMPSIFGAAGGGLRL 258
Query: 57 NENNLNGNNNTCPLYVLQEKLEVKNGQAVTFTPYNAKKGVILTSTDLNIKSYVTKTTCAQ 116
+ N+ CPL VLQ+ EV NG V FT A +I T+T L+I ++ TK CA+
Sbjct: 259 GKTK----NSKCPLTVLQDYSEVVNGLPVKFTRLEAGHDIISTNTALDI-AFTTKPDCAE 423
Query: 117 SQVWKL----NKVLSGVWFLATGGVEGNPGFDTIFNWFKIEKADKDYVFSFCPSVCKCQT 172
S W L NK L+G W + GG E N G I +FKI+K Y FCP +
Sbjct: 424 SSKWVLVDDFNK-LTGPW-VGIGGTEDNEGKHIINGFFKIQKHGFGYKLVFCPDITAPPG 597
Query: 173 LCRELGLYVYDHGKKHLALSDQVPSFRVVF 202
C ++G + D + L L++ P + VVF
Sbjct: 598 ACYDIGRH-DDFTGRLLVLANNDP-YEVVF 681
>TC82725 weakly similar to PIR|T07871|T07871 miraculin homolog root-knot
nematode-induced - tomato, partial (51%)
Length = 818
Score = 87.0 bits (214), Expect(2) = 7e-19
Identities = 65/186 (34%), Positives = 92/186 (48%), Gaps = 11/186 (5%)
Frame = +2
Query: 1 MKTSFLAFSIIFLAFICKTFAAP--EPVLDISGKKVTTGVKYYIL---PVIRGK---GGG 52
MK++FLAF ++ +A + + E V+DI+GK + Y +L P + G G
Sbjct: 59 MKSTFLAF-LLLIALTSQPLLSSSLEHVVDITGKNLRANAYYNVLLSMPYTNSRSPEGLG 235
Query: 53 LTVVNENNLNGNNNTCPLYVLQEKLEVKNGQAVTFTPYNAKKGVILTSTDLNIKSYVTKT 112
L+ N CPL V+ + + FTP N KKGVI S+DLNI +
Sbjct: 236 LS-------NNIGQPCPLDVIV--VSRYQSLPIRFTPLNLKKGVIRVSSDLNIMFRSNSS 388
Query: 113 TCAQSQVWKLNK--VLSGVWFLATGGVEGNPGFDTIFNWFKIEKADKDYVFSFCPSVC-K 169
+ VWKL++ G F+ T G GNPG +I NWFKIEK + Y +CP VC
Sbjct: 389 CPYHTTVWKLDRFDASKGKSFVTTDGFIGNPGPQSISNWFKIEKYVEGYKLVYCPIVCPS 568
Query: 170 CQTLCR 175
C+ C+
Sbjct: 569 CKHECK 586
Score = 23.1 bits (48), Expect(2) = 7e-19
Identities = 13/28 (46%), Positives = 21/28 (74%)
Frame = +3
Query: 177 LGLYVYDHGKKHLALSDQVPSFRVVFKR 204
+GL+ ++G K LALSD VP ++V F++
Sbjct: 591 VGLFEDENGNKRLALSD-VP-YQVKFRK 668
>TC88358 proteinase inhibitor 20 [Medicago truncatula]
Length = 806
Score = 89.4 bits (220), Expect = 9e-19
Identities = 68/203 (33%), Positives = 97/203 (47%), Gaps = 6/203 (2%)
Frame = +2
Query: 6 LAFSIIFLAFICKTFAAP----EPVLDISGKKVTTGVKYYILPVIRGKGGGLTVVNENNL 61
L S FI A E VLDI+G + G +YYILP +RG GGG +
Sbjct: 56 LTLSFFIFVFITNPSLATSNDVEQVLDINGNPIFPGGQYYILPALRGPGGGGVRLGRT-- 229
Query: 62 NGNNNTCPLYVLQEKLEVKNGQAVTFTPYNAKKGVILTSTDLNIKSYVTKTTCAQSQVWK 121
+ CP+ VLQ++ EVKNG V FT G+I T T L I+ Y K +CA S W
Sbjct: 230 --GDLKCPVTVLQDRREVKNGLPVKFTIPGISPGIIFTGTPLEIE-YTKKPSCAASTKWL 400
Query: 122 L--NKVLSGVWFLATGGVEGNPGFDTIFNWFKIEKADKDYVFSFCPSVCKCQTLCRELGL 179
+ + V+ G + GG E PG T+ F I+K + ++ V T C ++G
Sbjct: 401 IFVDNVI-GKACIGIGGPENYPGVQTLKGKFNIQKHASGFGYNLGFCVTGSPT-CLDIGR 574
Query: 180 YVYDHGKKHLALSDQVPSFRVVF 202
+ D + L L++ ++VVF
Sbjct: 575 FDNDEAGRRLNLTEH-EVYQVVF 640
>TC76747 similar to GP|20269067|emb|CAD29731. protease inhibitor {Sesbania
rostrata}, partial (86%)
Length = 1189
Score = 87.8 bits (216), Expect = 3e-18
Identities = 66/193 (34%), Positives = 99/193 (51%), Gaps = 8/193 (4%)
Frame = +3
Query: 21 AAPEPVLDISGKKVTTGVKYYILPVIRGKGGGLTVVNENNLNGNNNTCPLYVLQEKLEVK 80
A PE V+D G + G YY+ P + GG+T+ + N TCPL V ++
Sbjct: 171 AEPEAVVDKQGNPLKPGEGYYVFP-LWADNGGITLGHTRN-----KTCPLDV------IR 314
Query: 81 NGQAVTFTPYNAKKGV--ILTSTDLNIKSYVTKTTCAQSQVWKLNKVLSGVWFLATGGVE 138
N A+ Y + G+ I T TDL I+ + + C + +VW+L KV SG WF++TGG
Sbjct: 315 NPDAIGTPVYFSASGLDYIPTLTDLTIEIPILGSPCNEPKVWRLLKVGSGFWFVSTGGAA 494
Query: 139 GNPGFDTIFNWFKIEK-----ADKDYVFSFCPSVCKCQTLCRELGLYVYDHGKKHLALSD 193
G+ + + FKIE+ A + Y F FCPSV LC +G +V G K +A+ D
Sbjct: 495 GD-----LVSKFKIERLAGEHAYEIYSFKFCPSV--PGVLCAPVGTFVDTDGTKVMAVGD 653
Query: 194 QVPS-FRVVFKRA 205
+ + V F++A
Sbjct: 654 GIEEPYYVRFQKA 692
>TC80652 similar to GP|14161088|emb|CAB76907. alpha-fucosidase {Cicer
arietinum}, partial (81%)
Length = 788
Score = 87.0 bits (214), Expect = 4e-18
Identities = 73/212 (34%), Positives = 103/212 (48%), Gaps = 15/212 (7%)
Frame = +2
Query: 6 LAFSIIFLAFICKTFA-------APEPVLDISGKKVTTGVKYYILPVIRGK-GGGLTVVN 57
L+ ++ FL F+ T A E VLDI+G + G KYYILP IRG GGGL +
Sbjct: 11 LSLTLSFLLFVFTTNLSLAFSNDAVEQVLDINGNPIFPGGKYYILPAIRGPLGGGLRLG- 187
Query: 58 ENNLNGNNNTCPLYVLQEKLEVKNGQAVTFTPYNAKKGVILTSTDLNIKSYVTKTTCAQS 117
+N+ C + V+Q+ EV NG V F+ G+I T T ++I+ + K C +S
Sbjct: 188 ----KSSNSDCEVTVVQDYNEVINGVPVKFSIPEISPGIIFTGTPIDIE-FTKKPNCVES 352
Query: 118 QVWKL--NKVLSGVWFLATGGVEGNPGFDTIFNWFKIEKADK--DYVFSFCPSVCKCQTL 173
W + + V+ + GG E PGF T+ F IEK + Y +C K
Sbjct: 353 SKWLIFVDSVIQKA-CVGIGGPENYPGFRTLSGTFNIEKHESGFGYRLGYC---VKDSPT 520
Query: 174 CRELGL---YVYDHGKKHLALSDQVPSFRVVF 202
C ++G V D G L L+ QV +F VVF
Sbjct: 521 CLDIGRAHEEVEDEGGSRLHLTHQV-AFAVVF 613
>TC85685 similar to GP|14161088|emb|CAB76907. alpha-fucosidase {Cicer
arietinum}, partial (42%)
Length = 870
Score = 81.3 bits (199), Expect = 2e-16
Identities = 71/206 (34%), Positives = 101/206 (48%), Gaps = 4/206 (1%)
Frame = +1
Query: 1 MKTSFLAFSIIFLAFICKTFAAPEPVLDISGKKVTTGVKYYILPVI-RGKGGGLTVVNEN 59
MK++ FS++ +F A E V DI+G V G KYYI P+I +G GGGL +
Sbjct: 55 MKSTLFTFSLLLFSFTYFPLAFTETVEDINGNPVFPGGKYYIAPLISKGGGGGLKLGKT- 231
Query: 60 NLNGNNNTCPLYVLQEKLEVKNGQAVTFTPYNAKKGVILTSTDLNIKSYVTKTTCAQSQV 119
++ CP+ VLQ+ EV G V FT K+GVI T+ +++I+ +V K CA+S
Sbjct: 232 ----GDSECPVTVLQDFSEVVRGLPVRFT-IIVKRGVIFTTDEVDIE-FVKKPKCAESAK 393
Query: 120 WKLNKVLSGVWFLATGGVEGNPGFDTIFNWFKIE---KADKDYVFSFCPSVCKCQTLCRE 176
W VL+ F T V D FKIE Y +CP C +
Sbjct: 394 W----VLAHDDF-PTSWVGIGDNIDAFQGKFKIETLGSGSGAYKLVYCPLFSAPPGACSD 558
Query: 177 LGLYVYDHGKKHLALSDQVPSFRVVF 202
+G Y ++G + L ++ P FRVVF
Sbjct: 559 IGRYRDENGWR-LVPTENDP-FRVVF 630
>TC78632 similar to GP|9367042|gb|AAF87095.1| trypsin inhibitor {Glycine
max}, partial (44%)
Length = 803
Score = 78.2 bits (191), Expect = 2e-15
Identities = 68/208 (32%), Positives = 101/208 (47%), Gaps = 5/208 (2%)
Frame = +2
Query: 1 MKTSFLAFSIIFLAFICKTF---AAPEPVLDISGKKVTTGVKYYILPVIRGKGGGLTVVN 57
MK +FL ++ L F T+ A E V D +G + ++Y+ P I G GG +
Sbjct: 8 MKPTFLT-TLSLLLFALATYFPLAFTEQVRDSNGNPIFFSSRFYVKPSIFGAAGGGVKLG 184
Query: 58 ENNLNGNNNTCPLYVLQEKLEVKNGQAVTFTPYNAKKGVILTSTDLNIKSYV--TKTTCA 115
E N++CPL VLQ+ EV NG V F+ +A+ + L STD + V K CA
Sbjct: 185 ETG----NSSCPLTVLQDYSEVVNGLPVKFST-DAEIFIDLISTDTSRVDIVFPEKPECA 349
Query: 116 QSQVWKLNKVLSGVWFLATGGVEGNPGFDTIFNWFKIEKADKDYVFSFCPSVCKCQTLCR 175
+S W L + ++ GG+E G I FKI K Y FCP+ LC
Sbjct: 350 ESSKWLLIEDDFPRPWVGIGGIEDYIGKHIIDGKFKIVKHGFGYKLVFCPTFTAPPGLCH 529
Query: 176 ELGLYVYDHGKKHLALSDQVPSFRVVFK 203
++G Y +G++ L L++ P + VVF+
Sbjct: 530 DIGRYDDKNGRR-LILTEDDP-YEVVFE 607
>TC76750 similar to GP|20269067|emb|CAD29731. protease inhibitor {Sesbania
rostrata}, partial (77%)
Length = 651
Score = 76.3 bits (186), Expect = 8e-15
Identities = 54/165 (32%), Positives = 84/165 (50%), Gaps = 7/165 (4%)
Frame = +3
Query: 23 PEPVLDISGKKVTTGVKYYILPVIRGKGGGLTVVNENNLNGNNNTCPLYVLQEKLEVKNG 82
PE V+D G + G YY+ P + GG+T+ N TCPL V++ + G
Sbjct: 72 PEAVVDKQGNPLKPGEGYYVFP-LWADNGGITLGQTRN-----KTCPLDVIRNPEAI--G 227
Query: 83 QAVTFTPYNAKKGVILTSTDLNIKSYVTKTTCAQSQVWKLNK--VLSGVWFLATGGVEGN 140
V F Y + I T TDL ++ + + C++ +VWK++K + WF++TGG GN
Sbjct: 228 SPVYF--YEYEHDYIPTLTDLTVEIPILGSPCSERKVWKISKEGTRARFWFVSTGGFPGN 401
Query: 141 PGFDTIFNWFKIEKADKD-----YVFSFCPSVCKCQTLCRELGLY 180
+F+ FKIE+ + + Y F +CPSV TLC +G +
Sbjct: 402 -----LFSQFKIERLEGEHAYEIYSFLYCPSV--PGTLCAPVGTF 515
>TC86085
Length = 884
Score = 73.6 bits (179), Expect = 5e-14
Identities = 63/210 (30%), Positives = 103/210 (49%), Gaps = 9/210 (4%)
Frame = +3
Query: 4 SFLAFSIIFLAFICKTFAAPEPVLDISGKKVTTGVKYYILPVIRGKGGGLTVVNENNLNG 63
S F + + + T + + V+D SG+ V +Y+I P I G GGG T+V
Sbjct: 72 SLTIFILAHVWLLMATTSIAQFVIDTSGEPVEDDEEYFIRPAITGNGGGSTLVT------ 233
Query: 64 NNNTCPLYVLQEKLEVKNGQAVTFTPYNAK--KGVILTSTDLNIKSYVTKTTCAQSQVWK 121
N CPL+V + E G AV FTP+ + + + DL + +++T T+C QS W+
Sbjct: 234 GNGPCPLHVGLDNTEGTLGVAVKFTPFAPQHDDDDVRLNRDLRV-TFLTSTSCGQSTDWR 410
Query: 122 LNK--VLSGVWFLATGGVEGNPGFDTIFNWFKIEKADK--DYVFSFCPSVC--KCQTLCR 175
L + SG + TG + G + N+F+I + Y +CP+ C+ C
Sbjct: 411 LGEKDATSGRRLIVTG---RDNGAGSQGNFFRIVQTQTGGTYNIQWCPTEACPSCKVQCG 581
Query: 176 ELGLYVYDHGKKHLAL-SDQVPSFRVVFKR 204
+G+ + ++GK LAL D +P VVF++
Sbjct: 582 TVGV-IRENGKNLLALDGDALP---VVFQK 659
>BF636459
Length = 519
Score = 72.8 bits (177), Expect = 9e-14
Identities = 52/168 (30%), Positives = 81/168 (47%), Gaps = 4/168 (2%)
Frame = +1
Query: 3 TSFLAFSIIFLAFICKTFAAPEPVLDISGKKVTTGVKYYILPVIRGKGGGLTVVNENNLN 62
TS +F++ + E +LD G + G +YYI P I GG T+++ N
Sbjct: 49 TSLSLMLCLFMSIKTLAQSENEKILDTKGHPLERGKEYYIKPAITDSGGRFTLIDRNG-- 222
Query: 63 GNNNTCPLYVLQEKLEVKNGQAVTFTPYNAKKGVILTSTDLNIKSYVTKTTCAQSQVWKL 122
+CPLYV QE ++ G V FTP+ + VI S D +K + + C QS WKL
Sbjct: 223 ----SCPLYVGQENTDLGKGLPVIFTPFAKEDKVIKDSRDFKVK-FSASSICVQSTEWKL 387
Query: 123 --NKVLSGVWFLATGGVEGNPGFDTIFNWFKIEKADKDYVFS--FCPS 166
SG + + G+ G ++F+I KA+ + V++ FCP+
Sbjct: 388 GDRDTKSGRRVI----IAGSDG-----SYFRIVKAEFEGVYNIRFCPT 504
>TC89443 similar to GP|7208767|emb|CAB76906.1 alpha-fucosidase {Cicer
arietinum}, partial (78%)
Length = 735
Score = 65.1 bits (157), Expect = 2e-11
Identities = 69/208 (33%), Positives = 90/208 (43%), Gaps = 11/208 (5%)
Frame = +3
Query: 6 LAFSIIFLAFICKTFA--APEPVLDISGKKVTTGVKYYILPVIRG-KGGGLTVVNENNLN 62
L S AFI A + VLDI G + G +YYI P GGLT+ +L
Sbjct: 48 LTISFFLFAFITNLSPNNAAKQVLDIHGTPLIPGSQYYIFPASENPNSGGLTLNKVGDLE 227
Query: 63 GNNNTCPLYVLQEKLEVKNGQAVTFTPYNAKKGVILTSTDLNIKSYVTKTTCAQSQVWKL 122
CP+ VLQ + G V FT G ILT TDL I+ + K CA+S W +
Sbjct: 228 -----CPVTVLQNNAMI--GLPVKFTVPENNTGNILTGTDLEIE-FTKKPDCAESSKWLM 383
Query: 123 ----NKVLSGVWFLATGGVEGNPGFDTIFNWFKIEKADKDYVF--SFCPSVCKCQTLCRE 176
N LS V GG G +TI F I K +V+ FC V C
Sbjct: 384 FLDHNTQLSCV---GIGGATNYHGIETISGKFLIVKHGSGHVYRLGFCLDVTGD---CGY 545
Query: 177 LGLYVY--DHGKKHLALSDQVPSFRVVF 202
+GL ++ + G L L+ V ++ VVF
Sbjct: 546 IGLQMFNSEEGGSRLFLT-AVDAYSVVF 626
>TC82454 weakly similar to GP|3551243|dbj|BAA32820.1 Water-Soluble
Chlorophyll Protein {Brassica oleracea}, partial (17%)
Length = 940
Score = 64.3 bits (155), Expect = 3e-11
Identities = 63/212 (29%), Positives = 87/212 (40%), Gaps = 10/212 (4%)
Frame = +2
Query: 1 MKTSFLAFSIIFLAFICKTF------AAPEPVLDISGKKVTTGVKYYILPVIRGKGGGLT 54
MK + LA ++ FL F T+ E V D +G + KY+I P G GGGL
Sbjct: 38 MKPTLLA-TLPFLLFALTTYFPLPFTQGVEQVKDKNGNPILVSKKYFIWPA-DGSGGGLR 211
Query: 55 VVNENNLNGNNNTCPLYVLQEKLEVKNGQAVTFTPYNAKKGVILTS-TDLNIKSYVTKTT 113
+ CPL V Q E + F P I T T L+I + KT
Sbjct: 212 L-------NETEQCPLVVQQAFSEDVKSLPLKFIPTENINDFIFTGYTSLDIV-FEKKTK 367
Query: 114 CAQSQVWKLNKV-LSGVWFLATGGVEGNPGFDTIFNWFKIE--KADKDYVFSFCPSVCKC 170
CA+S W + K W GGV G D +F KIE ++ + Y FCP++
Sbjct: 368 CAESSKWVVVKGGFMEPWIGIGGGVNGKSVIDGLF---KIETIRSFRGYKLVFCPTISDP 538
Query: 171 QTLCRELGLYVYDHGKKHLALSDQVPSFRVVF 202
C +G + + L +S+ F VVF
Sbjct: 539 TGQCNNIGRFFDNENGLRLIMSENFKPFEVVF 634
>TC78015 similar to GP|12721693|gb|AAK03411.1 RbsC {Pasteurella multocida},
partial (4%)
Length = 776
Score = 63.2 bits (152), Expect = 7e-11
Identities = 59/194 (30%), Positives = 94/194 (48%), Gaps = 11/194 (5%)
Frame = -3
Query: 9 SIIFLAFIC---KTFAAPEPVLDISGKKVTTGVKYYILPVIRGKGGGLTVVNENNLNGNN 65
++I LA +C T + VLD G+ V +Y+I PVI KGG T+V+ N
Sbjct: 729 TLIILAHVCLFITTTTIAQFVLDTVGEPVEGDEEYFIRPVITNKGGRSTMVS------RN 568
Query: 66 NTCPLYVLQEKLEVKNGQAVTFTPYNAKKGV--ILTSTDLNIKSYVTKTTCAQSQVWKLN 123
+CPL+V E + G V FTP+ + + DL I ++ ++C QS W+L
Sbjct: 567 ESCPLHVGLELTGLGRGLVVKFTPFAPHHDFDDVRVNRDLRI-TFQASSSCVQSTEWRLG 391
Query: 124 K--VLSGVWFLATGGVEGNPGFDTIFNWFKIEKADKD--YVFSFCPS-VC-KCQTLCREL 177
+ SG + TG G + N+F+I + + Y +CP+ VC C+ C +
Sbjct: 390 EKDTKSGRRLIITGTDSATNG--SYGNFFRIVETPLEGMYNIQWCPTEVCPSCKFECGTV 217
Query: 178 GLYVYDHGKKHLAL 191
+ + ++GK LAL
Sbjct: 216 DM-LNENGKILLAL 178
>TC86612 weakly similar to GP|7208767|emb|CAB76906.1 alpha-fucosidase {Cicer
arietinum}, partial (81%)
Length = 808
Score = 62.0 bits (149), Expect = 2e-10
Identities = 58/189 (30%), Positives = 83/189 (43%), Gaps = 8/189 (4%)
Frame = +3
Query: 6 LAFSIIFLAFICKTFAAP----EPVLDISGKKVTTGVKYYILPVI-RGKGGGLTVVNENN 60
L+ ++ F F T +P +PV D G +T G +YYILP GGLT+
Sbjct: 54 LSLTLSFFLFTFITNLSPNNAVQPVFDKHGNPLTPGNQYYILPASDNPSSGGLTLDKV-- 227
Query: 61 LNGNNNTCPLYVLQEKLEVKNGQAVTFTPYNAKKGVILTSTDLNIKSYVTKTTCAQSQVW 120
++ CPL VLQ G V FT I+T TDL I+ ++ K CA+S W
Sbjct: 228 ---GDSVCPLTVLQN--NAVTGLPVKFTILENSTSNIVTGTDLEIE-FINKPDCAESSKW 389
Query: 121 -KLNKVLSGVWFLATGGVEGNPGFDTIFNWFKIEK--ADKDYVFSFCPSVCKCQTLCREL 177
+ ++ + F+ GG PG + I F I K + Y FC C L
Sbjct: 390 LMVVDHVTQLSFVGIGGPANYPGVELISGKFLILKHGSGNAYRVGFCLDTTGD---CAYL 560
Query: 178 GLYVYDHGK 186
GL ++ G+
Sbjct: 561 GLQEFNSGE 587
>TC86086 similar to GP|15155442|gb|AAK86324.1 AGR_C_899p {Agrobacterium
tumefaciens str. C58 (Cereon)}, partial (2%)
Length = 1329
Score = 59.7 bits (143), Expect = 8e-10
Identities = 56/182 (30%), Positives = 82/182 (44%), Gaps = 9/182 (4%)
Frame = -3
Query: 19 TFAAPEPVLDISGKKVTTGVKYYILPVIRGKGGGLTVVNENNLNGNNNTCPLYVLQEKLE 78
T + + V+D SG+ V +Y+I P I G GGG +V N CPL+V E
Sbjct: 1207 TTSVAQIVIDTSGEPVEDDEEYFIRPAITGNGGGSILVT------RNGPCPLHVGLGNSE 1046
Query: 79 VKNGQAVTFTPYNAKKGVILTSTDLNIKSYVT---KTTCAQSQVWKLNK--VLSGVWFLA 133
G AV FTP+ + LN VT T C QS W+L + SG +
Sbjct: 1045 GTLGLAVKFTPFAPRHDDDDDDVRLNRDLRVTFQGFTGCGQSTDWRLGEKDATSGRRLIV 866
Query: 134 TGGVEGNPGFDTIFNWFKIEKADKD--YVFSFCPSVC--KCQTLCRELGLYVYDHGKKHL 189
TG + G + N+F+I + Y +CP+ C+ C +G+ + ++GK L
Sbjct: 865 TG---RDNGAGSHGNFFRIVQTQTGGIYNIQWCPTEACPSCKVQCGTVGV-IRENGKILL 698
Query: 190 AL 191
AL
Sbjct: 697 AL 692
>AJ500060
Length = 345
Score = 49.7 bits (117), Expect = 8e-07
Identities = 29/88 (32%), Positives = 43/88 (47%)
Frame = +2
Query: 4 SFLAFSIIFLAFICKTFAAPEPVLDISGKKVTTGVKYYILPVIRGKGGGLTVVNENNLNG 63
S F + + + T + + V+D SG+ V +Y+I P I G GGG T++
Sbjct: 98 SLTIFILAHVWLLMATKSIAQFVIDTSGEPVEDDEEYFIRPAITGNGGGFTLIT------ 259
Query: 64 NNNTCPLYVLQEKLEVKNGQAVTFTPYN 91
N TCPL V + E G VTF P++
Sbjct: 260 GNGTCPLNVGLDNTEGTLGAPVTFIPFS 343
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.322 0.139 0.426
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,189,936
Number of Sequences: 36976
Number of extensions: 107188
Number of successful extensions: 601
Number of sequences better than 10.0: 52
Number of HSP's better than 10.0 without gapping: 570
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 571
length of query: 205
length of database: 9,014,727
effective HSP length: 92
effective length of query: 113
effective length of database: 5,612,935
effective search space: 634261655
effective search space used: 634261655
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 56 (26.2 bits)
Medicago: description of AC140022.4


