

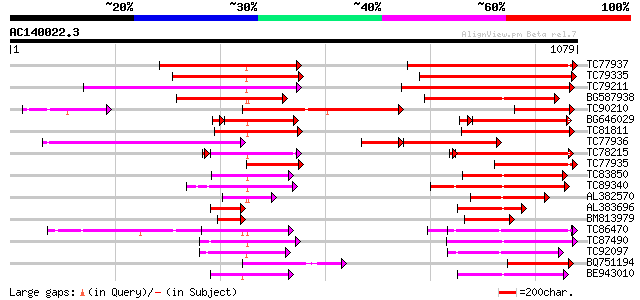
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140022.3 - phase: 0
(1079 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC77937 similar to GP|14715462|dbj|BAB62040. CjMDR1 {Coptis japo... 427 e-119
TC79335 similar to GP|14715462|dbj|BAB62040. CjMDR1 {Coptis japo... 407 e-113
TC79211 similar to GP|4204793|gb|AAD10836.1| P-glycoprotein {Sol... 333 2e-91
BG587938 homologue to GP|22655312|g putative ABC transporter {Ar... 315 5e-86
TC90210 similar to GP|14715462|dbj|BAB62040. CjMDR1 {Coptis japo... 305 8e-83
BG646029 similar to GP|14715462|d CjMDR1 {Coptis japonica}, part... 265 1e-75
TC81811 similar to GP|6671365|gb|AAF23176.1| P-glycoprotein {Gos... 271 1e-72
TC77936 weakly similar to PIR|T52319|T52319 P-glycoprotein-like ... 242 5e-64
TC78215 similar to GP|10177549|dbj|BAB10828. ABC transporter-lik... 230 2e-60
TC77935 similar to PIR|E85023|E85023 probable P-glycoprotein-lik... 221 9e-58
TC83850 weakly similar to GP|7300071|gb|AAF55241.1| CG4225 gene ... 201 1e-51
TC89340 weakly similar to GP|17427488|emb|CAD14006. PROBABLE COM... 201 1e-51
AL382570 weakly similar to GP|10241621|em putative P-glycoprotei... 178 9e-45
AL383696 homologue to PIR|T47671|T4 P-glycoprotein-like - Arabid... 148 1e-35
BM813979 homologue to PIR|E85023|E85 probable P-glycoprotein-lik... 143 3e-34
TC86470 similar to PIR|F84919|F84919 glutathione-conjugate trans... 143 3e-34
TC87490 similar to PIR|T52080|T52080 multi resistance protein [i... 132 9e-31
TC92097 similar to GP|22553016|emb|CAD44995. multidrug-resistanc... 130 3e-30
BQ751194 similar to GP|21622336|em probable multidrug resistance... 130 4e-30
BE943010 similar to GP|6634763|gb Similar to gb|AF008124 Arabido... 127 2e-29
>TC77937 similar to GP|14715462|dbj|BAB62040. CjMDR1 {Coptis japonica},
partial (24%)
Length = 1164
Score = 427 bits (1098), Expect = e-119
Identities = 217/322 (67%), Positives = 266/322 (82%)
Frame = +3
Query: 758 FYIGSILVHHRKATFVEIFRVFFSLTMTAMSVSQSSTLFPDTNKAIDSAASIFNILDSKP 817
FY G+ LV K++F ++FRVFF+L+M A+ +SQS +L PD+ KA + ASIF ILD K
Sbjct: 15 FYAGARLVEDGKSSFSDVFRVFFALSMAAIGLSQSGSLLPDSTKAKSAVASIFAILDRKS 194
Query: 818 DIDSSSNDGVTQETVVGNIELQHVNFSYPTRPDIQIFKDLTLSIPSAKTVALVGESGSGK 877
ID + G+T E V G IE +HVNF YPTRPDIQIF+DL L+I S KTVALVGESGSGK
Sbjct: 195 LIDPTDESGITLEEVKGEIEFKHVNFKYPTRPDIQIFRDLCLNIHSGKTVALVGESGSGK 374
Query: 878 STVISLLERFYDPNSGRVLLDGVDIKTFRISWLRQQMGLVGQEPILFNESIRANIAYGKE 937
STVISL++RFYDP+SG + LDG +I++ ++ WLRQQMGLV QEP+LFN++IRANIAYGK
Sbjct: 375 STVISLIQRFYDPDSGHITLDGKEIQSLQVKWLRQQMGLVSQEPVLFNDTIRANIAYGKG 554
Query: 938 DGATEDEIIAAANAANAHNFISSLPDGYDTSVGERGTQLSGGQKQRIAIARAMLKNPKIL 997
A+E EIIAAA ANAH FISSL GYDT VGERG QLSGGQKQR+AIARA++KNPKIL
Sbjct: 555 GDASEAEIIAAAELANAHKFISSLQKGYDTVVGERGVQLSGGQKQRVAIARAIVKNPKIL 734
Query: 998 LLDEATSALDAESERIVQEALDRVSLNRTTVIVAHRLTTIRGADTIAVIKNGMVAEKGRH 1057
LLDEATSALDAESE++VQ+ALDRV + RTT+IVAHRL+TI+GAD IAV+KNG++AEKG+H
Sbjct: 735 LLDEATSALDAESEKVVQDALDRVMVERTTIIVAHRLSTIKGADLIAVVKNGVIAEKGKH 914
Query: 1058 DELMNNTHGVYASLVALHSTAS 1079
+ L++ G YASLVALH++AS
Sbjct: 915 EALLHK-GGDYASLVALHTSAS 977
Score = 249 bits (636), Expect = 4e-66
Identities = 140/318 (44%), Positives = 194/318 (60%), Gaps = 49/318 (15%)
Frame = +3
Query: 286 AMWYGSKLVIAKGYNGGTVMTVVIALVTGSMSLGQTSPSLHAFAAGKAAAYKMFETIKRK 345
+ + G++LV + V V AL ++ L Q+ L K+A +F + RK
Sbjct: 12 SFYAGARLVEDGKSSFSDVFRVFFALSMAAIGLSQSGSLLPDSTKAKSAVASIFAILDRK 191
Query: 346 PKIDAYDTSGLVLEDIKGDIELRDVHFRYPARPDVEIFAGFSLFVPSGTTTALVGQSGSG 405
ID D SG+ LE++KG+IE + V+F+YP RPD++IF L + SG T ALVG+SGSG
Sbjct: 192 SLIDPTDESGITLEEVKGEIEFKHVNFKYPTRPDIQIFRDLCLNIHSGKTVALVGESGSG 371
Query: 406 KSTVISLLERFYDPNAGEVLIDGVNLKNLQLRWIREQIGLVSQE---------------- 449
KSTVISL++RFYDP++G + +DG +++LQ++W+R+Q+GLVSQE
Sbjct: 372 KSTVISLIQRFYDPDSGHITLDGKEIQSLQVKWLRQQMGLVSQEPVLFNDTIRANIAYGK 551
Query: 450 ---------------------------------RQNGTQLSGGQKQRIAIARAILKNPKI 476
+ G QLSGGQKQR+AIARAI+KNPKI
Sbjct: 552 GGDASEAEIIAAAELANAHKFISSLQKGYDTVVGERGVQLSGGQKQRVAIARAIVKNPKI 731
Query: 477 LLLDEATSALDAESEHIVQEALEKIILKRTTIVVAHRLTTIIHADTIAVVQQGKIVERGT 536
LLLDEATSALDAESE +VQ+AL++++++RTTI+VAHRL+TI AD IAVV+ G I E+G
Sbjct: 732 LLLDEATSALDAESEKVVQDALDRVMVERTTIIVAHRLSTIKGADLIAVVKNGVIAEKGK 911
Query: 537 HSELTMDPHGAYSQLIRL 554
H E + G Y+ L+ L
Sbjct: 912 H-EALLHKGGDYASLVAL 962
>TC79335 similar to GP|14715462|dbj|BAB62040. CjMDR1 {Coptis japonica},
partial (23%)
Length = 1135
Score = 407 bits (1046), Expect = e-113
Identities = 201/299 (67%), Positives = 249/299 (83%)
Frame = +2
Query: 780 FSLTMTAMSVSQSSTLFPDTNKAIDSAASIFNILDSKPDIDSSSNDGVTQETVVGNIELQ 839
F+LTM A+ +SQSS+ PD++KA + ASIF ++D K ID S G T +++ G IEL+
Sbjct: 2 FALTMAAIGISQSSSFAPDSSKAKSATASIFGMIDKKSKIDPSEESGTTLDSIKGEIELR 181
Query: 840 HVNFSYPTRPDIQIFKDLTLSIPSAKTVALVGESGSGKSTVISLLERFYDPNSGRVLLDG 899
H++F YP+RPDIQIF+DL L+I S KTVALVGESGSGKSTVI+LL+RFYDP+SG + LDG
Sbjct: 182 HISFKYPSRPDIQIFRDLNLTIHSGKTVALVGESGSGKSTVIALLQRFYDPDSGEITLDG 361
Query: 900 VDIKTFRISWLRQQMGLVGQEPILFNESIRANIAYGKEDGATEDEIIAAANAANAHNFIS 959
++I+ ++ WLRQQMGLV QEP+LFN++IRANIAYGK ATE EIIAAA ANAH FIS
Sbjct: 362 IEIRQLQLKWLRQQMGLVSQEPVLFNDTIRANIAYGKGGIATEAEIIAAAELANAHRFIS 541
Query: 960 SLPDGYDTSVGERGTQLSGGQKQRIAIARAMLKNPKILLLDEATSALDAESERIVQEALD 1019
L GYDT VGERGTQLSGGQKQR+AIARA++K+PKILLLDEATSALDAESER+VQ+ALD
Sbjct: 542 GLQQGYDTIVGERGTQLSGGQKQRVAIARAIIKSPKILLLDEATSALDAESERVVQDALD 721
Query: 1020 RVSLNRTTVIVAHRLTTIRGADTIAVIKNGMVAEKGRHDELMNNTHGVYASLVALHSTA 1078
+V +NRTTV+VAHRL+TI+ AD IAV+KNG++ EKGRH+ L+N G YASLV LH++A
Sbjct: 722 KVMVNRTTVVVAHRLSTIKNADVIAVVKNGVIVEKGRHETLINVKDGFYASLVQLHTSA 898
Score = 259 bits (663), Expect = 3e-69
Identities = 140/299 (46%), Positives = 190/299 (62%), Gaps = 49/299 (16%)
Frame = +2
Query: 310 ALVTGSMSLGQTSPSLHAFAAGKAAAYKMFETIKRKPKIDAYDTSGLVLEDIKGDIELRD 369
AL ++ + Q+S + K+A +F I +K KID + SG L+ IKG+IELR
Sbjct: 5 ALTMAAIGISQSSSFAPDSSKAKSATASIFGMIDKKSKIDPSEESGTTLDSIKGEIELRH 184
Query: 370 VHFRYPARPDVEIFAGFSLFVPSGTTTALVGQSGSGKSTVISLLERFYDPNAGEVLIDGV 429
+ F+YP+RPD++IF +L + SG T ALVG+SGSGKSTVI+LL+RFYDP++GE+ +DG+
Sbjct: 185 ISFKYPSRPDIQIFRDLNLTIHSGKTVALVGESGSGKSTVIALLQRFYDPDSGEITLDGI 364
Query: 430 NLKNLQLRWIREQIGLVSQE---------------------------------------- 449
++ LQL+W+R+Q+GLVSQE
Sbjct: 365 EIRQLQLKWLRQQMGLVSQEPVLFNDTIRANIAYGKGGIATEAEIIAAAELANAHRFISG 544
Query: 450 ---------RQNGTQLSGGQKQRIAIARAILKNPKILLLDEATSALDAESEHIVQEALEK 500
+ GTQLSGGQKQR+AIARAI+K+PKILLLDEATSALDAESE +VQ+AL+K
Sbjct: 545 LQQGYDTIVGERGTQLSGGQKQRVAIARAIIKSPKILLLDEATSALDAESERVVQDALDK 724
Query: 501 IILKRTTIVVAHRLTTIIHADTIAVVQQGKIVERGTHSELTMDPHGAYSQLIRLQEGEK 559
+++ RTT+VVAHRL+TI +AD IAVV+ G IVE+G H L G Y+ L++L K
Sbjct: 725 VMVNRTTVVVAHRLSTIKNADVIAVVKNGVIVEKGRHETLINVKDGFYASLVQLHTSAK 901
>TC79211 similar to GP|4204793|gb|AAD10836.1| P-glycoprotein {Solanum
tuberosum}, partial (36%)
Length = 1667
Score = 333 bits (854), Expect = 2e-91
Identities = 178/331 (53%), Positives = 235/331 (70%), Gaps = 2/331 (0%)
Frame = +1
Query: 746 AHSSLYCTNAFIFYIGSILVHHRKATFVEIFRVFFSLTMTAMSVSQSSTLFPDTNKAIDS 805
A +LY + A + S LV H + F + RVF L ++A +++ TL PD K +
Sbjct: 409 AQFALYASYALGLWYASWLVKHGISDFSKTIRVFMVLMVSANGAAETLTLAPDFIKGGRA 588
Query: 806 AASIFNILDSKPDIDSSSNDGV-TQETVVGNIELQHVNFSYPTRPDIQIFKDLTLSIPSA 864
S+F++LD + +I+ D + + G +EL+HV+FSYPTRPD+ +F+DL L I +
Sbjct: 589 MRSVFDLLDRQTEIEPDDQDATPVPDRLRGEVELKHVDFSYPTRPDMPVFRDLNLRIRAG 768
Query: 865 KTVALVGESGSGKSTVISLLERFYDPNSGRVLLDGVDIKTFRISWLRQQMGLVGQEPILF 924
KT+ALVG SG GKS+VI+L++RFYDP SGR+++DG DI+ + + LR+ + +V QEP LF
Sbjct: 769 KTLALVGPSGCGKSSVIALIQRFYDPTSGRIMIDGKDIRKYNLKSLRRHISVVPQEPCLF 948
Query: 925 NESIRANIAYGKEDGATEDEIIAAANAANAHNFISSLPDGYDTSVGERGTQLSGGQKQRI 984
+I NIAYG D ATE EII AA ANAH FISSLPDGY T VGERG QLSGGQKQRI
Sbjct: 949 ATTIYENIAYG-HDSATEAEIIEAATLANAHKFISSLPDGYKTFVGERGVQLSGGQKQRI 1125
Query: 985 AIARAMLKNPKILLLDEATSALDAESERIVQEALDRVSLNRTTVIVAHRLTTIRGADTIA 1044
A+ARA L+ +++LLDEATSALDAESER VQEALDR S +TT+IVAHRL+TIR A+ IA
Sbjct: 1126 AVARAFLRKAELMLLDEATSALDAESERSVQEALDRASTGKTTIIVAHRLSTIRNANVIA 1305
Query: 1045 VIKNGMVAEKGRHDELM-NNTHGVYASLVAL 1074
VI +G VAE+G H +LM N+ G+YA ++ L
Sbjct: 1306 VIDDGKVAEQGSHSQLMKNHQDGIYARMIQL 1398
Score = 267 bits (682), Expect = 2e-71
Identities = 156/465 (33%), Positives = 249/465 (53%), Gaps = 50/465 (10%)
Frame = +1
Query: 141 TGEVISRMSGDTILIQEAMGEKVGKFLQLGSTFFGGFVIAFIKGWRLALVLLACVPCIVV 200
+ + +R++ D ++ A+G+++ +Q + F+ WRLALVL+A P +V
Sbjct: 7 SARISARLALDANNVRSAIGDRISVIVQNTALMLVACTAGFVLQWRLALVLIAVFPVVVA 186
Query: 201 AGAFMAMVMAKMAIRGQVAYAEAGNVANQTVGSMRTVASFTGEKKAIEKYNSKIKIAYTA 260
A M M + + A+A+A +A + + ++RTVA+F E K + + S ++
Sbjct: 187 ATVLQKMFMTGFSGDLEAAHAKATQLAGEAIANVRTVAAFNSESKIVRLFASNLETPLQR 366
Query: 261 MVQQSIASGIGMGTLLLIIFCSYGLAMWYGSKLVIAKGYNGGTVMTVVIALVTGSMSLGQ 320
+ SG G G ++ SY L +WY S LV + + V + L+ + +
Sbjct: 367 CFWKGQISGSGYGIAQFALYASYALGLWYASWLVKHGISDFSKTIRVFMVLMVSANGAAE 546
Query: 321 TSPSLHAFAAGKAAAYKMFETIKRKPKIDAYDTSGLVLED-IKGDIELRDVHFRYPARPD 379
T F G A +F+ + R+ +I+ D + D ++G++EL+ V F YP RPD
Sbjct: 547 TLTLAPDFIKGGRAMRSVFDLLDRQTEIEPDDQDATPVPDRLRGEVELKHVDFSYPTRPD 726
Query: 380 VEIFAGFSLFVPSGTTTALVGQSGSGKSTVISLLERFYDPNAGEVLIDGVNLKNLQLRWI 439
+ +F +L + +G T ALVG SG GKS+VI+L++RFYDP +G ++IDG +++ L+ +
Sbjct: 727 MPVFRDLNLRIRAGKTLALVGPSGCGKSSVIALIQRFYDPTSGRIMIDGKDIRKYNLKSL 906
Query: 440 REQIGLVSQE------------------------------------------------RQ 451
R I +V QE +
Sbjct: 907 RRHISVVPQEPCLFATTIYENIAYGHDSATEAEIIEAATLANAHKFISSLPDGYKTFVGE 1086
Query: 452 NGTQLSGGQKQRIAIARAILKNPKILLLDEATSALDAESEHIVQEALEKIILKRTTIVVA 511
G QLSGGQKQRIA+ARA L+ +++LLDEATSALDAESE VQEAL++ +TTI+VA
Sbjct: 1087RGVQLSGGQKQRIAVARAFLRKAELMLLDEATSALDAESERSVQEALDRASTGKTTIIVA 1266
Query: 512 HRLTTIIHADTIAVVQQGKIVERGTHSELTMD-PHGAYSQLIRLQ 555
HRL+TI +A+ IAV+ GK+ E+G+HS+L + G Y+++I+LQ
Sbjct: 1267HRLSTIRNANVIAVIDDGKVAEQGSHSQLMKNHQDGIYARMIQLQ 1401
>BG587938 homologue to GP|22655312|g putative ABC transporter {Arabidopsis
thaliana}, partial (19%)
Length = 780
Score = 315 bits (808), Expect = 5e-86
Identities = 161/258 (62%), Positives = 202/258 (77%)
Frame = +3
Query: 789 VSQSSTLFPDTNKAIDSAASIFNILDSKPDIDSSSNDGVTQETVVGNIELQHVNFSYPTR 848
+ QS+ KA +AA IF I+D +P ID +S G+ ETV G +EL++V+FSYP+R
Sbjct: 3 LGQSAPSMAAFTKARVAAAKIFRIIDHQPGIDRNSESGLELETVTGLVELKNVDFSYPSR 182
Query: 849 PDIQIFKDLTLSIPSAKTVALVGESGSGKSTVISLLERFYDPNSGRVLLDGVDIKTFRIS 908
P++ I D +LS+P+ KT+ALVG SGSGKSTV+SL+ERFYDP SG+V+LDG DIKT ++
Sbjct: 183 PEVLILNDFSLSVPAGKTIALVGSSGSGKSTVVSLIERFYDPTSGQVMLDGHDIKTLKLK 362
Query: 909 WLRQQMGLVGQEPILFNESIRANIAYGKEDGATEDEIIAAANAANAHNFISSLPDGYDTS 968
WLRQQ+GLV QEP LF +IR NI G+ D A + EI AA ANAH+FI LP+G++T
Sbjct: 363 WLRQQIGLVSQEPALFATTIRENILLGRPD-ANQVEIEEAARVANAHSFIIKLPEGFETQ 539
Query: 969 VGERGTQLSGGQKQRIAIARAMLKNPKILLLDEATSALDAESERIVQEALDRVSLNRTTV 1028
VGERG QLSGGQKQRIAIARAMLKNP ILLLDEATSALD+ESE++VQEALDR + RTT+
Sbjct: 540 VGERGLQLSGGQKQRIAIARAMLKNPAILLLDEATSALDSESEKLVQEALDRFMIGRTTL 719
Query: 1029 IVAHRLTTIRGADTIAVI 1046
++AHRL+TIR AD +AVI
Sbjct: 720 VIAHRLSTIRKADLVAVI 773
Score = 236 bits (601), Expect = 5e-62
Identities = 131/259 (50%), Positives = 170/259 (65%), Gaps = 48/259 (18%)
Frame = +3
Query: 318 LGQTSPSLHAFAAGKAAAYKMFETIKRKPKIDAYDTSGLVLEDIKGDIELRDVHFRYPAR 377
LGQ++PS+ AF + AA K+F I +P ID SGL LE + G +EL++V F YP+R
Sbjct: 3 LGQSAPSMAAFTKARVAAAKIFRIIDHQPGIDRNSESGLELETVTGLVELKNVDFSYPSR 182
Query: 378 PDVEIFAGFSLFVPSGTTTALVGQSGSGKSTVISLLERFYDPNAGEVLIDGVNLKNLQLR 437
P+V I FSL VP+G T ALVG SGSGKSTV+SL+ERFYDP +G+V++DG ++K L+L+
Sbjct: 183 PEVLILNDFSLSVPAGKTIALVGSSGSGKSTVVSLIERFYDPTSGQVMLDGHDIKTLKLK 362
Query: 438 WIREQIGLVSQE--------RQN------------------------------------- 452
W+R+QIGLVSQE R+N
Sbjct: 363 WLRQQIGLVSQEPALFATTIRENILLGRPDANQVEIEEAARVANAHSFIIKLPEGFETQV 542
Query: 453 ---GTQLSGGQKQRIAIARAILKNPKILLLDEATSALDAESEHIVQEALEKIILKRTTIV 509
G QLSGGQKQRIAIARA+LKNP ILLLDEATSALD+ESE +VQEAL++ ++ RTT+V
Sbjct: 543 GERGLQLSGGQKQRIAIARAMLKNPAILLLDEATSALDSESEKLVQEALDRFMIGRTTLV 722
Query: 510 VAHRLTTIIHADTIAVVQQ 528
+AHRL+TI AD +AV+ +
Sbjct: 723 IAHRLSTIRKADLVAVIHK 779
>TC90210 similar to GP|14715462|dbj|BAB62040. CjMDR1 {Coptis japonica},
partial (26%)
Length = 1158
Score = 305 bits (780), Expect = 8e-83
Identities = 162/310 (52%), Positives = 217/310 (69%), Gaps = 4/310 (1%)
Frame = +2
Query: 444 GLVSQERQNGTQLSGGQKQRIAIARAILKNPKILLLDEATSALDAESEHIVQEALEKIIL 503
GL + +GTQLSGGQKQRIAIARAILKNP+ILLLDEATSALDAESE +VQEAL++I++
Sbjct: 8 GLDTMVGDHGTQLSGGQKQRIAIARAILKNPRILLLDEATSALDAESERVVQEALDRIMV 187
Query: 504 KRTTIVVAHRLTTIIHADTIAVVQQGKIVERGTHSELTMDPHGAYSQLIRLQEGEKEAEG 563
RTT+VVAHRL+T+ +AD IAV+ +GK+VE+GTHSEL DP GAYSQLIRLQE KE+E
Sbjct: 188 NRTTVVVAHRLSTVRNADMIAVIHRGKMVEKGTHSELLKDPEGAYSQLIRLQEVNKESE- 364
Query: 564 SRSSEVDKFGDNLNIDIHMAGSSTQRISFVRSISQTSSMS----HRHSQLSGEIVDANIE 619
E + S+QR S RSIS+ SS+ H S G N+
Sbjct: 365 ----ETTDHHGKRELSAESFRQSSQRKSLQRSISRGSSIGNSSRHSFSVSFGLPTGVNVA 532
Query: 620 QGQVDNNEKPKMSMKNSIWRLAKLNKPELPVILLGTIAAMVNGVVFPIFGFLFSAVISMF 679
++ + + + RLA LNKPE+PV+L+G++AA+ NGV+ PIFG L S+VI F
Sbjct: 533 DPDLEKVPTKEKEQEVPLRRLASLNKPEIPVLLIGSLAAIANGVILPIFGVLISSVIKTF 712
Query: 680 YKPPEQQRKESRFWSLVYVGLGLVTLVVFPLKNYFFGTAGGKLIERIRSLTFAKIVHQEI 739
Y+P ++ +K+S+FW+++++ LGL +LVV P + YFF AG KLI+RIR L F K+V+ E+
Sbjct: 713 YEPFDEMKKDSKFWAIMFMLLGLASLVVIPARGYFFSVAGCKLIQRIRLLCFEKVVNMEV 892
Query: 740 RWFDDPAHSS 749
W+D+P +SS
Sbjct: 893 GWYDEPENSS 922
Score = 162 bits (411), Expect = 5e-40
Identities = 80/113 (70%), Positives = 94/113 (82%)
Frame = +2
Query: 962 PDGYDTSVGERGTQLSGGQKQRIAIARAMLKNPKILLLDEATSALDAESERIVQEALDRV 1021
P G DT VG+ GTQLSGGQKQRIAIARA+LKNP+ILLLDEATSALDAESER+VQEALDR+
Sbjct: 2 PQGLDTMVGDHGTQLSGGQKQRIAIARAILKNPRILLLDEATSALDAESERVVQEALDRI 181
Query: 1022 SLNRTTVIVAHRLTTIRGADTIAVIKNGMVAEKGRHDELMNNTHGVYASLVAL 1074
+NRTTV+VAHRL+T+R AD IAVI G + EKG H EL+ + G Y+ L+ L
Sbjct: 182 MVNRTTVVVAHRLSTVRNADMIAVIHRGKMVEKGTHSELLKDPEGAYSQLIRL 340
Score = 79.0 bits (193), Expect = 9e-15
Identities = 48/176 (27%), Positives = 97/176 (54%), Gaps = 5/176 (2%)
Frame = +2
Query: 24 QIVPFYKLFSFADRLDVTLMIIGTISAMANGFASPLMTLLLGKVINAFGSSNQSEVLNQV 83
Q VP +L S ++ ++ +++IG+++A+ANG P+ +L+ VI F E +++
Sbjct: 572 QEVPLRRLASL-NKPEIPVLLIGSLAAIANGVILPIFGVLISSVIKTF-----YEPFDEM 733
Query: 84 SKVSLLFVYLAIGSGITSFLQVSC----WMVTGERQSARIRSLYLKTILKQDIAFFDT-E 138
K S + + + G+ S + + + V G + RIR L + ++ ++ ++D E
Sbjct: 734 KKDSKFWAIMFMLLGLASLVVIPARGYFFSVAGCKLIQRIRLLCFEKVVNMEVGWYDEPE 913
Query: 139 TNTGEVISRMSGDTILIQEAMGEKVGKFLQLGSTFFGGFVIAFIKGWRLALVLLAC 194
++G V +R+S D ++ +G+ +G +Q ++ G +IAFI W+LAL++L C
Sbjct: 914 NSSGAVGARLSADAASVRALVGDALGLLVQNLASALAGLIIAFIASWQLALIILVC 1081
>BG646029 similar to GP|14715462|d CjMDR1 {Coptis japonica}, partial (16%)
Length = 680
Score = 265 bits (677), Expect(2) = 1e-75
Identities = 130/189 (68%), Positives = 162/189 (84%)
Frame = -3
Query: 881 ISLLERFYDPNSGRVLLDGVDIKTFRISWLRQQMGLVGQEPILFNESIRANIAYGKEDGA 940
ISLL+RFYDP+SG++ LDG +I+ ++ W RQQMGLV QEP+LFN++IRANIAYGK A
Sbjct: 606 ISLLQRFYDPDSGQIKLDGTEIQKLQLKWFRQQMGLVSQEPVLFNDTIRANIAYGKGGNA 427
Query: 941 TEDEIIAAANAANAHNFISSLPDGYDTSVGERGTQLSGGQKQRIAIARAMLKNPKILLLD 1000
TE E+IAAA ANAHNFISSL GYDT VGERG QLSGGQKQ +AIARA++ P+ILLLD
Sbjct: 426 TEAEVIAAAELANAHNFISSLQQGYDTIVGERGIQLSGGQKQPVAIARAIVNRPRILLLD 247
Query: 1001 EATSALDAESERIVQEALDRVSLNRTTVIVAHRLTTIRGADTIAVIKNGMVAEKGRHDEL 1060
EATSALDAESE++VQ+ALDRV ++RTT++VAHRL+TI+GA++IAV+KNG++ EKG+HD L
Sbjct: 246 EATSALDAESEKVVQDALDRVRVDRTTIVVAHRLSTIKGANSIAVVKNGVIEEKGKHDIL 67
Query: 1061 MNNTHGVYA 1069
+N G YA
Sbjct: 66 INK-GGTYA 43
Score = 146 bits (368), Expect(2) = 4e-39
Identities = 84/189 (44%), Positives = 115/189 (60%), Gaps = 49/189 (25%)
Frame = -3
Query: 410 ISLLERFYDPNAGEVLIDGVNLKNLQLRWIREQIGLVSQER------------------- 450
ISLL+RFYDP++G++ +DG ++ LQL+W R+Q+GLVSQE
Sbjct: 606 ISLLQRFYDPDSGQIKLDGTEIQKLQLKWFRQQMGLVSQEPVLFNDTIRANIAYGKGGNA 427
Query: 451 ------------------------------QNGTQLSGGQKQRIAIARAILKNPKILLLD 480
+ G QLSGGQKQ +AIARAI+ P+ILLLD
Sbjct: 426 TEAEVIAAAELANAHNFISSLQQGYDTIVGERGIQLSGGQKQPVAIARAIVNRPRILLLD 247
Query: 481 EATSALDAESEHIVQEALEKIILKRTTIVVAHRLTTIIHADTIAVVQQGKIVERGTHSEL 540
EATSALDAESE +VQ+AL+++ + RTTIVVAHRL+TI A++IAVV+ G I E+G H ++
Sbjct: 246 EATSALDAESEKVVQDALDRVRVDRTTIVVAHRLSTIKGANSIAVVKNGVIEEKGKH-DI 70
Query: 541 TMDPHGAYS 549
++ G Y+
Sbjct: 69 LINKGGTYA 43
Score = 38.1 bits (87), Expect(2) = 1e-75
Identities = 19/24 (79%), Positives = 22/24 (91%)
Frame = -2
Query: 857 LTLSIPSAKTVALVGESGSGKSTV 880
L+L+I S +TVALVGESGSGKSTV
Sbjct: 679 LSLTIHSGQTVALVGESGSGKSTV 608
Score = 35.0 bits (79), Expect(2) = 4e-39
Identities = 17/23 (73%), Positives = 19/23 (81%)
Frame = -2
Query: 387 SLFVPSGTTTALVGQSGSGKSTV 409
SL + SG T ALVG+SGSGKSTV
Sbjct: 676 SLTIHSGQTVALVGESGSGKSTV 608
>TC81811 similar to GP|6671365|gb|AAF23176.1| P-glycoprotein {Gossypium
hirsutum}, partial (17%)
Length = 850
Score = 271 bits (692), Expect = 1e-72
Identities = 140/214 (65%), Positives = 170/214 (79%)
Frame = +1
Query: 861 IPSAKTVALVGESGSGKSTVISLLERFYDPNSGRVLLDGVDIKTFRISWLRQQMGLVGQE 920
+PS K+VALVG+SGSGKS+VISL+ RFYDP SG+VL+DG DI + L + +GLV QE
Sbjct: 1 VPSGKSVALVGQSGSGKSSVISLILRFYDPTSGKVLIDGKDITRINLKSLTKHIGLVQQE 180
Query: 921 PILFNESIRANIAYGKEDGATEDEIIAAANAANAHNFISSLPDGYDTSVGERGTQLSGGQ 980
P LF SI NI YGKE GA++ E+I AA ANAHNFIS+LP+GY T VGERG QLSGGQ
Sbjct: 181 PALFATSIYENILYGKE-GASDSEVIEAAKLANAHNFISALPEGYSTKVGERGVQLSGGQ 357
Query: 981 KQRIAIARAMLKNPKILLLDEATSALDAESERIVQEALDRVSLNRTTVIVAHRLTTIRGA 1040
+QR+AIARA+LKNP+ILLLDEATSALD ESERIVQ+ALDR+ NRTTV+VAHRL+TIR A
Sbjct: 358 RQRVAIARAVLKNPEILLLDEATSALDVESERIVQQALDRLMQNRTTVMVAHRLSTIRNA 537
Query: 1041 DTIAVIKNGMVAEKGRHDELMNNTHGVYASLVAL 1074
D I+V+++G + E+G H L+ N G Y LV L
Sbjct: 538 DQISVLQDGKIIEQGTHSSLIENKDGPYYKLVNL 639
Score = 187 bits (474), Expect = 2e-47
Identities = 104/215 (48%), Positives = 137/215 (63%), Gaps = 48/215 (22%)
Frame = +1
Query: 390 VPSGTTTALVGQSGSGKSTVISLLERFYDPNAGEVLIDGVNLKNLQLRWIREQIGLVSQE 449
VPSG + ALVGQSGSGKS+VISL+ RFYDP +G+VLIDG ++ + L+ + + IGLV QE
Sbjct: 1 VPSGKSVALVGQSGSGKSSVISLILRFYDPTSGKVLIDGKDITRINLKSLTKHIGLVQQE 180
Query: 450 R------------------------------------------------QNGTQLSGGQK 461
+ G QLSGGQ+
Sbjct: 181 PALFATSIYENILYGKEGASDSEVIEAAKLANAHNFISALPEGYSTKVGERGVQLSGGQR 360
Query: 462 QRIAIARAILKNPKILLLDEATSALDAESEHIVQEALEKIILKRTTIVVAHRLTTIIHAD 521
QR+AIARA+LKNP+ILLLDEATSALD ESE IVQ+AL++++ RTT++VAHRL+TI +AD
Sbjct: 361 QRVAIARAVLKNPEILLLDEATSALDVESERIVQQALDRLMQNRTTVMVAHRLSTIRNAD 540
Query: 522 TIAVVQQGKIVERGTHSELTMDPHGAYSQLIRLQE 556
I+V+Q GKI+E+GTHS L + G Y +L+ LQ+
Sbjct: 541 QISVLQDGKIIEQGTHSSLIENKDGPYYKLVNLQQ 645
>TC77936 weakly similar to PIR|T52319|T52319 P-glycoprotein-like protein
pgp3 [imported] - Arabidopsis thaliana, partial (32%)
Length = 1219
Score = 242 bits (618), Expect = 5e-64
Identities = 139/390 (35%), Positives = 223/390 (56%), Gaps = 2/390 (0%)
Frame = +1
Query: 62 LLLGKVINAFGSSNQSEVLNQVSKV-SLLFVYLAIGSGITSFLQVSCWMVTGERQSARIR 120
LL+ K+IN F + L SKV +++FV +A+ + + + + V G + RIR
Sbjct: 4 LLISKMINIF--YKPAHELRHDSKVWAIVFVAVAVATLLIIPCRFYFFGVAGGKLIQRIR 177
Query: 121 SLYLKTILKQDIAFFD-TETNTGEVISRMSGDTILIQEAMGEKVGKFLQLGSTFFGGFVI 179
++ + ++ ++++FD E ++G + +R+S D ++ +G+ +G +Q +T G VI
Sbjct: 178 NMCFEKVVHMEVSWFDEAEHSSGALGARLSTDAASVRALVGDALGLLVQNIATAIAGLVI 357
Query: 180 AFIKGWRLALVLLACVPCIVVAGAFMAMVMAKMAIRGQVAYAEAGNVANQTVGSMRTVAS 239
+F W+LA ++LA P + + G V+ + + Y EA VAN VGS+RTVAS
Sbjct: 358 SFQASWQLAFIVLALAPLLGLNGYVQVKVLKGFSADAKKLYEEASQVANDAVGSIRTVAS 537
Query: 240 FTGEKKAIEKYNSKIKIAYTAMVQQSIASGIGMGTLLLIIFCSYGLAMWYGSKLVIAKGY 299
F EKK +E Y K + V++ I SG G G +++ + G++LV
Sbjct: 538 FCAEKKVMELYKQKCEGPIKKGVRRGIISGFGFGLSFFMLYAVDACVFYAGARLVEDGKS 717
Query: 300 NGGTVMTVVIALVTGSMSLGQTSPSLHAFAAGKAAAYKMFETIKRKPKIDAYDTSGLVLE 359
V V AL +M + Q+ + K+AA +F + +ID+ D SG+ LE
Sbjct: 718 TFSDVFLVFFALTMAAMGVSQSGTLVPDSTNAKSAAASIFAILDHNSQIDSSDESGMTLE 897
Query: 360 DIKGDIELRDVHFRYPARPDVEIFAGFSLFVPSGTTTALVGQSGSGKSTVISLLERFYDP 419
++KGDIE V F+YP DV+IF L + SG T ALVG+SGSGKSTVISLL+RFYDP
Sbjct: 898 EVKGDIEFNHVSFKYPTTLDVQIFNDLCLNIRSGKTVALVGESGSGKSTVISLLQRFYDP 1077
Query: 420 NAGEVLIDGVNLKNLQLRWIREQIGLVSQE 449
++G + +DG+ ++ +Q++W+R+Q+GLVSQE
Sbjct: 1078DSGHITLDGIEIQRMQVKWLRQQMGLVSQE 1167
Score = 231 bits (589), Expect = 1e-60
Identities = 116/187 (62%), Positives = 141/187 (75%)
Frame = +1
Query: 750 LYCTNAFIFYIGSILVHHRKATFVEIFRVFFSLTMTAMSVSQSSTLFPDTNKAIDSAASI 809
LY +A +FY G+ LV K+TF ++F VFF+LTM AM VSQS TL PD+ A +AASI
Sbjct: 655 LYAVDACVFYAGARLVEDGKSTFSDVFLVFFALTMAAMGVSQSGTLVPDSTNAKSAAASI 834
Query: 810 FNILDSKPDIDSSSNDGVTQETVVGNIELQHVNFSYPTRPDIQIFKDLTLSIPSAKTVAL 869
F ILD IDSS G+T E V G+IE HV+F YPT D+QIF DL L+I S KTVAL
Sbjct: 835 FAILDHNSQIDSSDESGMTLEEVKGDIEFNHVSFKYPTTLDVQIFNDLCLNIRSGKTVAL 1014
Query: 870 VGESGSGKSTVISLLERFYDPNSGRVLLDGVDIKTFRISWLRQQMGLVGQEPILFNESIR 929
VGESGSGKSTVISLL+RFYDP+SG + LDG++I+ ++ WLRQQMGLV QEPILFN+++
Sbjct: 1015VGESGSGKSTVISLLQRFYDPDSGHITLDGIEIQRMQVKWLRQQMGLVSQEPILFNDTVX 1194
Query: 930 ANIAYGK 936
ANIAY K
Sbjct: 1195ANIAYXK 1215
Score = 98.2 bits (243), Expect = 2e-20
Identities = 40/81 (49%), Positives = 60/81 (73%)
Frame = +1
Query: 669 GFLFSAVISMFYKPPEQQRKESRFWSLVYVGLGLVTLVVFPLKNYFFGTAGGKLIERIRS 728
G L S +I++FYKP + R +S+ W++V+V + + TL++ P + YFFG AGGKLI+RIR+
Sbjct: 1 GLLISKMINIFYKPAHELRHDSKVWAIVFVAVAVATLLIIPCRFYFFGVAGGKLIQRIRN 180
Query: 729 LTFAKIVHQEIRWFDDPAHSS 749
+ F K+VH E+ WFD+ HSS
Sbjct: 181 MCFEKVVHMEVSWFDEAEHSS 243
>TC78215 similar to GP|10177549|dbj|BAB10828. ABC transporter-like protein
{Arabidopsis thaliana}, partial (36%)
Length = 1176
Score = 230 bits (586), Expect(2) = 2e-60
Identities = 116/224 (51%), Positives = 158/224 (69%)
Frame = +1
Query: 849 PDIQIFKDLTLSIPSAKTVALVGESGSGKSTVISLLERFYDPNSGRVLLDGVDIKTFRIS 908
P + K + + + VALVG SG GK+T+ +L+ERFYDP G +L++GV +
Sbjct: 34 P*YMVLKGINIKLQPGSKVALVGPSGGGKTTIANLIERFYDPTKGNILVNGVPLVEISHK 213
Query: 909 WLRQQMGLVGQEPILFNESIRANIAYGKEDGATEDEIIAAANAANAHNFISSLPDGYDTS 968
L +++ +V QEP LFN SI NIAYG + + +I AA ANAH FIS+ P+ Y T
Sbjct: 214 HLHRKISIVSQEPTLFNCSIEENIAYGFDGKIEDADIENAAKMANAHEFISNFPEKYKTF 393
Query: 969 VGERGTQLSGGQKQRIAIARAMLKNPKILLLDEATSALDAESERIVQEALDRVSLNRTTV 1028
VGERG +LSGGQKQRIAIARA+L +PKILLLDEATSALDAESE +VQ+A+D + RT +
Sbjct: 394 VGERGIRLSGGQKQRIAIARALLMDPKILLLDEATSALDAESEYLVQDAMDSIMKGRTVL 573
Query: 1029 IVAHRLTTIRGADTIAVIKNGMVAEKGRHDELMNNTHGVYASLV 1072
++AHRL+T++ A+T+AV+ +G + E G HDEL+ +GVY +LV
Sbjct: 574 VIAHRLSTVKTANTVAVVSDGQIVESGTHDELLEK-NGVYTALV 702
Score = 163 bits (412), Expect(2) = 1e-40
Identities = 92/223 (41%), Positives = 132/223 (58%), Gaps = 49/223 (21%)
Frame = +1
Query: 382 IFAGFSLFVPSGTTTALVGQSGSGKSTVISLLERFYDPNAGEVLIDGVNLKNLQLRWIRE 441
+ G ++ + G+ ALVG SG GK+T+ +L+ERFYDP G +L++GV L + + +
Sbjct: 46 VLKGINIKLQPGSKVALVGPSGGGKTTIANLIERFYDPTKGNILVNGVPLVEISHKHLHR 225
Query: 442 QIGLVSQER-------------------------------------------------QN 452
+I +VSQE +
Sbjct: 226 KISIVSQEPTLFNCSIEENIAYGFDGKIEDADIENAAKMANAHEFISNFPEKYKTFVGER 405
Query: 453 GTQLSGGQKQRIAIARAILKNPKILLLDEATSALDAESEHIVQEALEKIILKRTTIVVAH 512
G +LSGGQKQRIAIARA+L +PKILLLDEATSALDAESE++VQ+A++ I+ RT +V+AH
Sbjct: 406 GIRLSGGQKQRIAIARALLMDPKILLLDEATSALDAESEYLVQDAMDSIMKGRTVLVIAH 585
Query: 513 RLTTIIHADTIAVVQQGKIVERGTHSELTMDPHGAYSQLIRLQ 555
RL+T+ A+T+AVV G+IVE GTH EL ++ +G Y+ L++ Q
Sbjct: 586 RLSTVKTANTVAVVSDGQIVESGTHDEL-LEKNGVYTALVKRQ 711
Score = 23.1 bits (48), Expect(2) = 1e-40
Identities = 8/14 (57%), Positives = 11/14 (78%)
Frame = +2
Query: 367 LRDVHFRYPARPDV 380
L DV F YP+RP++
Sbjct: 2 LNDVWFSYPSRPNI 43
Score = 21.9 bits (45), Expect(2) = 2e-60
Identities = 9/14 (64%), Positives = 11/14 (78%)
Frame = +2
Query: 838 LQHVNFSYPTRPDI 851
L V FSYP+RP+I
Sbjct: 2 LNDVWFSYPSRPNI 43
>TC77935 similar to PIR|E85023|E85023 probable P-glycoprotein-like protein
[imported] - Arabidopsis thaliana, partial (12%)
Length = 869
Score = 221 bits (564), Expect = 9e-58
Identities = 113/157 (71%), Positives = 136/157 (85%)
Frame = +2
Query: 923 LFNESIRANIAYGKEDGATEDEIIAAANAANAHNFISSLPDGYDTSVGERGTQLSGGQKQ 982
LFN+++RANIAYGK ATE EI+AAA ANAH FI SL GYDT VGERG QLSGGQKQ
Sbjct: 65 LFNDTVRANIAYGKGGDATEAEIVAAAELANAHQFIGSLQKGYDTIVGERGIQLSGGQKQ 244
Query: 983 RIAIARAMLKNPKILLLDEATSALDAESERIVQEALDRVSLNRTTVIVAHRLTTIRGADT 1042
R+AIARA++KNPKILLLDEATSALDAESE++VQ+ALDRV + RTT+IVAHRL+TI+GAD
Sbjct: 245 RVAIARAIVKNPKILLLDEATSALDAESEKVVQDALDRVMVERTTIIVAHRLSTIKGADL 424
Query: 1043 IAVIKNGMVAEKGRHDELMNNTHGVYASLVALHSTAS 1079
IAV+KNG++AEKG+H+ L++ G YASLVALH++ S
Sbjct: 425 IAVVKNGVIAEKGKHEALLHK-GGDYASLVALHTSDS 532
Score = 129 bits (325), Expect = 5e-30
Identities = 67/108 (62%), Positives = 86/108 (79%)
Frame = +2
Query: 451 QNGTQLSGGQKQRIAIARAILKNPKILLLDEATSALDAESEHIVQEALEKIILKRTTIVV 510
+ G QLSGGQKQR+AIARAI+KNPKILLLDEATSALDAESE +VQ+AL++++++RTTI+V
Sbjct: 209 ERGIQLSGGQKQRVAIARAIVKNPKILLLDEATSALDAESEKVVQDALDRVMVERTTIIV 388
Query: 511 AHRLTTIIHADTIAVVQQGKIVERGTHSELTMDPHGAYSQLIRLQEGE 558
AHRL+TI AD IAVV+ G I E+G H E + G Y+ L+ L +
Sbjct: 389 AHRLSTIKGADLIAVVKNGVIAEKGKH-EALLHKGGDYASLVALHTSD 529
>TC83850 weakly similar to GP|7300071|gb|AAF55241.1| CG4225 gene product
{Drosophila melanogaster}, partial (23%)
Length = 816
Score = 201 bits (512), Expect = 1e-51
Identities = 103/200 (51%), Positives = 143/200 (71%)
Frame = +2
Query: 862 PSAKTVALVGESGSGKSTVISLLERFYDPNSGRVLLDGVDIKTFRISWLRQQMGLVGQEP 921
P KT A+VGESGSGKST + LL RFYD G V +DG DIK ++ LR+ +G+V Q+
Sbjct: 53 PGTKT-AIVGESGSGKSTCLKLLFRFYDVQEGAVTVDGHDIKHLKLDSLRRNIGVVPQDT 229
Query: 922 ILFNESIRANIAYGKEDGATEDEIIAAANAANAHNFISSLPDGYDTSVGERGTQLSGGQK 981
+LFN +I N+ Y A++ ++ A AAN H I + PDGY+T VGERG +LSGG++
Sbjct: 230 VLFNATIMYNLLYANPK-ASQADVFEACKAANIHERILNFPDGYETKVGERGLKLSGGER 406
Query: 982 QRIAIARAMLKNPKILLLDEATSALDAESERIVQEALDRVSLNRTTVIVAHRLTTIRGAD 1041
QRIAIARA+LK+ +ILLLDEAT++LD+ +ER +Q+AL+RV+ RTT+ +AHRL+TI +D
Sbjct: 407 QRIAIARAILKDARILLLDEATASLDSHTERQIQDALERVTAGRTTITIAHRLSTITTSD 586
Query: 1042 TIAVIKNGMVAEKGRHDELM 1061
I V+ + E+G H EL+
Sbjct: 587 QIVVLHKSKIVERGTHSELL 646
Score = 152 bits (383), Expect = 9e-37
Identities = 89/204 (43%), Positives = 119/204 (57%), Gaps = 48/204 (23%)
Frame = +2
Query: 385 GFSLFVPSGTTTALVGQSGSGKSTVISLLERFYDPNAGEVLIDGVNLKNLQLRWIREQIG 444
G S V GT TA+VG+SGSGKST + LL RFYD G V +DG ++K+L+L +R IG
Sbjct: 32 GVSFTVAPGTKTAIVGESGSGKSTCLKLLFRFYDVQEGAVTVDGHDIKHLKLDSLRRNIG 211
Query: 445 LVSQER------------------------------------------------QNGTQL 456
+V Q+ + G +L
Sbjct: 212 VVPQDTVLFNATIMYNLLYANPKASQADVFEACKAANIHERILNFPDGYETKVGERGLKL 391
Query: 457 SGGQKQRIAIARAILKNPKILLLDEATSALDAESEHIVQEALEKIILKRTTIVVAHRLTT 516
SGG++QRIAIARAILK+ +ILLLDEAT++LD+ +E +Q+ALE++ RTTI +AHRL+T
Sbjct: 392 SGGERQRIAIARAILKDARILLLDEATASLDSHTERQIQDALERVTAGRTTITIAHRLST 571
Query: 517 IIHADTIAVVQQGKIVERGTHSEL 540
I +D I V+ + KIVERGTHSEL
Sbjct: 572 ITTSDQIVVLHKSKIVERGTHSEL 643
>TC89340 weakly similar to GP|17427488|emb|CAD14006. PROBABLE COMPOSITE
ATP-BINDING TRANSMEMBRANE ABC TRANSPORTER PROTEIN
{Ralstonia solanacearum}, partial (32%)
Length = 795
Score = 201 bits (512), Expect = 1e-51
Identities = 116/268 (43%), Positives = 164/268 (60%), Gaps = 3/268 (1%)
Frame = +1
Query: 801 KAIDSAASIFNILDSKPDIDSSSNDGVTQETVVGNIELQHVNFSYPTRPDIQIFKDLTLS 860
+A+ S + + KP + S ++ GNI ++V F+Y P + KDL
Sbjct: 4 QALISGERLLELFKVKPTVVDSPTAEPLRDCN-GNIRWKNVGFNYD--PKNKALKDLDFE 174
Query: 861 IPSAKTVALVGESGSGKSTVISLLERFYDPNSGRVLLDGVDIKTFRISWLRQQMGLVGQE 920
T A VGESG GKSTV L+ R+Y+ SG + +DG D+K I +R +G+V Q+
Sbjct: 175 CKPGSTTAFVGESGGGKSTVFRLMFRYYNTTSGSIEIDGRDVKDLTIDSVRSYIGVVPQD 354
Query: 921 PILFNESIRANIAYGKEDGATEDEIIAAANAANAHNFISSLPDGYDTSVGERGTQLSGGQ 980
LFNES+ N+ Y + + AT++EI A AA+ H+ I S PD YDT VGERG +LSGG+
Sbjct: 355 TTLFNESLMYNLRYARPE-ATDEEIYDACRAASIHDRIMSFPDRYDTKVGERGLKLSGGE 531
Query: 981 KQRIAIARAMLKNPKILLLDEATSALDAESERIVQEALDRVSLNRTTVIVAHRLTTIRGA 1040
+QR+AIAR ++K+PKI++LDEATSALDA +E+ +Q+ L + RT +I+AHRL+TI A
Sbjct: 532 RQRVAIARTIIKSPKIIMLDEATSALDAHTEQQIQDRLGHLGQGRTLLIIAHRLSTITHA 711
Query: 1041 DTIAVIKNGMVAEKGRHDEL---MNNTH 1065
D I V + KG H EL M+ TH
Sbjct: 712 DQIIVPQQWDY**KGHHKELVMPMDGTH 795
Score = 139 bits (349), Expect = 8e-33
Identities = 90/260 (34%), Positives = 131/260 (49%), Gaps = 48/260 (18%)
Frame = +1
Query: 337 KMFETIKRKPKIDAYDTSGLVLEDIKGDIELRDVHFRYPARPDVEIFAGFSLFVPSGTTT 396
++ E K KP + T+ L D G+I ++V F Y P + G+TT
Sbjct: 25 RLLELFKVKPTVVDSPTAE-PLRDCNGNIRWKNVGFNYD--PKNKALKDLDFECKPGSTT 195
Query: 397 ALVGQSGSGKSTVISLLERFYDPNAGEVLIDGVNLKNLQLRWIREQIGLVSQER------ 450
A VG+SG GKSTV L+ R+Y+ +G + IDG ++K+L + +R IG+V Q+
Sbjct: 196 AFVGESGGGKSTVFRLMFRYYNTTSGSIEIDGRDVKDLTIDSVRSYIGVVPQDTTLFNES 375
Query: 451 ------------------------------------------QNGTQLSGGQKQRIAIAR 468
+ G +LSGG++QR+AIAR
Sbjct: 376 LMYNLRYARPEATDEEIYDACRAASIHDRIMSFPDRYDTKVGERGLKLSGGERQRVAIAR 555
Query: 469 AILKNPKILLLDEATSALDAESEHIVQEALEKIILKRTTIVVAHRLTTIIHADTIAVVQQ 528
I+K+PKI++LDEATSALDA +E +Q+ L + RT +++AHRL+TI HAD I V QQ
Sbjct: 556 TIIKSPKIIMLDEATSALDAHTEQQIQDRLGHLGQGRTLLIIAHRLSTITHADQIIVPQQ 735
Query: 529 GKIVERGTHSELTMDPHGAY 548
+G H EL M G +
Sbjct: 736 WDY**KGHHKELVMPMDGTH 795
>AL382570 weakly similar to GP|10241621|em putative P-glycoprotein {Oryza
sativa}, partial (17%)
Length = 481
Score = 178 bits (452), Expect = 9e-45
Identities = 91/151 (60%), Positives = 115/151 (75%)
Frame = +3
Query: 877 KSTVISLLERFYDPNSGRVLLDGVDIKTFRISWLRQQMGLVGQEPILFNESIRANIAYGK 936
KS+V++LL +FYDP G+VL+DG D++ + + WLR Q+GLV QEP+LFN SIR NI YG
Sbjct: 3 KSSVLALLLKFYDPVVGKVLIDGKDLREYNLRWLRTQIGLVQQEPLLFNCSIRENICYGN 182
Query: 937 EDGATEDEIIAAANAANAHNFISSLPDGYDTSVGERGTQLSGGQKQRIAIARAMLKNPKI 996
+GA E EI+ A AN H F+S+LP+GY+T VGE+G QLSGGQKQRIAIAR +LK P I
Sbjct: 183 -NGAFESEIVEVAREANIHEFVSNLPNGYNTVVGEKGCQLSGGQKQRIAIARTLLKKPAI 359
Query: 997 LLLDEATSALDAESERIVQEALDRVSLNRTT 1027
LLLDEATSALDA SER + A+ ++L T
Sbjct: 360 LLLDEATSALDA*SERTIVNAIKAMNLKEET 452
Score = 101 bits (252), Expect = 1e-21
Identities = 66/150 (44%), Positives = 80/150 (53%), Gaps = 48/150 (32%)
Frame = +3
Query: 406 KSTVISLLERFYDPNAGEVLIDGVNLKNLQLRWIREQIGLVSQE--------RQN----- 452
KS+V++LL +FYDP G+VLIDG +L+ LRW+R QIGLV QE R+N
Sbjct: 3 KSSVLALLLKFYDPVVGKVLIDGKDLREYNLRWLRTQIGLVQQEPLLFNCSIRENICYGN 182
Query: 453 -----------------------------------GTQLSGGQKQRIAIARAILKNPKIL 477
G QLSGGQKQRIAIAR +LK P IL
Sbjct: 183 NGAFESEIVEVAREANIHEFVSNLPNGYNTVVGEKGCQLSGGQKQRIAIARTLLKKPAIL 362
Query: 478 LLDEATSALDAESEHIVQEALEKIILKRTT 507
LLDEATSALDA SE + A++ + LK T
Sbjct: 363 LLDEATSALDA*SERTIVNAIKAMNLKEET 452
>AL383696 homologue to PIR|T47671|T4 P-glycoprotein-like - Arabidopsis
thaliana, partial (9%)
Length = 391
Score = 148 bits (374), Expect = 1e-35
Identities = 73/130 (56%), Positives = 97/130 (74%)
Frame = +1
Query: 853 IFKDLTLSIPSAKTVALVGESGSGKSTVISLLERFYDPNSGRVLLDGVDIKTFRISWLRQ 912
+ + +L + +TVA+VG SGSGKST+ISL+ERFYDP +G+VLLDG D+K + + WLR
Sbjct: 4 VLSNFSLKVNGGQTVAVVGVSGSGKSTIISLIERFYDPVAGQVLLDGRDLKLYNLRWLRS 183
Query: 913 QMGLVGQEPILFNESIRANIAYGKEDGATEDEIIAAANAANAHNFISSLPDGYDTSVGER 972
+GL+ QEPI+F+ +IR NI Y + + A+E E+ AA ANAH+FISSLP GYDT VG R
Sbjct: 184 HLGLIQQEPIIFSTTIRENIIYARHN-ASEAEMKEAARIANAHHFISSLPHGYDTHVGMR 360
Query: 973 GTQLSGGQKQ 982
G L+ GQKQ
Sbjct: 361 GVDLTPGQKQ 390
Score = 86.7 bits (213), Expect = 5e-17
Identities = 41/68 (60%), Positives = 52/68 (76%)
Frame = +1
Query: 382 IFAGFSLFVPSGTTTALVGQSGSGKSTVISLLERFYDPNAGEVLIDGVNLKNLQLRWIRE 441
+ + FSL V G T A+VG SGSGKST+ISL+ERFYDP AG+VL+DG +LK LRW+R
Sbjct: 4 VLSNFSLKVNGGQTVAVVGVSGSGKSTIISLIERFYDPVAGQVLLDGRDLKLYNLRWLRS 183
Query: 442 QIGLVSQE 449
+GL+ QE
Sbjct: 184 HLGLIQQE 207
>BM813979 homologue to PIR|E85023|E85 probable P-glycoprotein-like protein
[imported] - Arabidopsis thaliana, partial (7%)
Length = 434
Score = 143 bits (361), Expect = 3e-34
Identities = 70/97 (72%), Positives = 83/97 (85%)
Frame = +3
Query: 865 KTVALVGESGSGKSTVISLLERFYDPNSGRVLLDGVDIKTFRISWLRQQMGLVGQEPILF 924
+TVALVGESGSGKSTVI+LL+RFYDP+SG + LDG++I+ ++ WLRQQMGLV QEP+LF
Sbjct: 135 QTVALVGESGSGKSTVIALLQRFYDPDSGEITLDGIEIRQLQLKWLRQQMGLVSQEPVLF 314
Query: 925 NESIRANIAYGKEDGATEDEIIAAANAANAHNFISSL 961
N++IRANIAYGK ATE EIIAAA ANAH FIS L
Sbjct: 315 NDTIRANIAYGKGGIATEAEIIAAAELANAHRFISGL 425
Score = 87.0 bits (214), Expect = 3e-17
Identities = 37/55 (67%), Positives = 51/55 (92%)
Frame = +3
Query: 395 TTALVGQSGSGKSTVISLLERFYDPNAGEVLIDGVNLKNLQLRWIREQIGLVSQE 449
T ALVG+SGSGKSTVI+LL+RFYDP++GE+ +DG+ ++ LQL+W+R+Q+GLVSQE
Sbjct: 138 TVALVGESGSGKSTVIALLQRFYDPDSGEITLDGIEIRQLQLKWLRQQMGLVSQE 302
>TC86470 similar to PIR|F84919|F84919 glutathione-conjugate transporter AtMRP4
[imported] - Arabidopsis thaliana, partial (57%)
Length = 3175
Score = 143 bits (361), Expect = 3e-34
Identities = 76/247 (30%), Positives = 143/247 (57%), Gaps = 1/247 (0%)
Frame = +1
Query: 834 GNIELQHVNFSYPTRPDIQ-IFKDLTLSIPSAKTVALVGESGSGKSTVISLLERFYDPNS 892
G+++++ + Y RP+ + K +TLSI + V +VG +GSGKST+I + R +P
Sbjct: 2017 GHVDIKDLQVRY--RPNTPLVLKGITLSISGGEKVGVVGRTGSGKSTLIQVFFRLVEPTG 2190
Query: 893 GRVLLDGVDIKTFRISWLRQQMGLVGQEPILFNESIRANIAYGKEDGATEDEIIAAANAA 952
G++++DG+DI + LR + G++ QEP+LF ++R+NI T+DEI + +
Sbjct: 2191 GKIIIDGIDICALGLHDLRSRFGIIPQEPVLFEGTVRSNI--DPTGQYTDDEIWKSLDRC 2364
Query: 953 NAHNFISSLPDGYDTSVGERGTQLSGGQKQRIAIARAMLKNPKILLLDEATSALDAESER 1012
+ ++S P+ D+ V + G S GQ+Q + + R MLK ++L +DEAT+++D++++
Sbjct: 2365 QLKDTVASKPEKLDSLVVDNGDNWSVGQRQLLCLGRVMLKQSRLLFMDEATASVDSQTDA 2544
Query: 1013 IVQEALDRVSLNRTTVIVAHRLTTIRGADTIAVIKNGMVAEKGRHDELMNNTHGVYASLV 1072
++Q+ + RT + +AHR+ T+ D + V+ G E + L+ ++A+LV
Sbjct: 2545 VIQKIIREDFAARTIISIAHRIPTVMDCDRVLVVDAGRAKEFDKPSNLLQR-QSLFAALV 2721
Query: 1073 ALHSTAS 1079
++ S
Sbjct: 2722 QEYANRS 2742
Score = 116 bits (291), Expect = 4e-26
Identities = 117/525 (22%), Positives = 226/525 (42%), Gaps = 57/525 (10%)
Frame = +1
Query: 73 SSNQSEVLNQVSKVSLLFVYLAIGSGITSFLQVSCWMVT--GERQSARIRSLYLKTILKQ 130
S ++EV N V +S +Y AI + V + VT G + + + L +IL
Sbjct: 1150 SVERAEVFNPVVFIS---IYAAITIVSVILIVVRSYSVTIFGLKTAQIFFNQILTSILHA 1320
Query: 131 DIAFFDTETNTGEVISRMSGDTILIQEAMGEKVGKFLQLGSTFFGGFVIAFIKGWRLALV 190
++F+DT T +G ++SR S D + + + + + T +I W A +
Sbjct: 1321 PMSFYDT-TPSGRILSRASTDQTNVDIFIPLFINFVVAMYITVISIVIITCQNSWPTAFL 1497
Query: 191 LLACVPCIVVAGAFMAMVMAKMAIRGQVAYAEAGNVANQTVGSMRTVASFTGEKKA---- 246
L+ V + + ++ + A ++++ + TV +F +K+
Sbjct: 1498 LIPLVWLNIWYRGYFLSTSRELTRLDSITKAPVIVHFSESISGVMTVRAFRKQKEFRLEN 1677
Query: 247 IEKYNSKIKIAYTAMVQQSIASGIGMGTLLLIIFCSYGLAMWYGSKLVIAKGYNGGTVMT 306
++ NS +++ + + G + L ++FC L M +I K N G ++
Sbjct: 1678 FKRVNSNLRMDFHNYSSNAWL-GFRLELLGSLVFCLSALFMILLPSNII-KPENVGLSLS 1851
Query: 307 VVIALVTGSMSLGQTSPSLHAFAAGKAAAY---KMFETIKRKPKIDAYDTSGLVLEDIKG 363
++L S+ + + F K + K F I + + D S +G
Sbjct: 1852 YGLSL----NSVLFWAIYMSCFIENKMVSVERIKQFSNIPSEAAWNIKDRSPPPNWPGQG 2019
Query: 364 DIELRDVHFRYPARPDVE-IFAGFSLFVPSGTTTALVGQSGSGKSTVISLLERFYDPNAG 422
++++D+ RY RP+ + G +L + G +VG++GSGKST+I + R +P G
Sbjct: 2020 HVDIKDLQVRY--RPNTPLVLKGITLSISGGEKVGVVGRTGSGKSTLIQVFFRLVEPTGG 2193
Query: 423 EVLIDGVNLKNLQLRWIREQIGLVSQER-------------------------------- 450
+++IDG+++ L L +R + G++ QE
Sbjct: 2194 KIIIDGIDICALGLHDLRSRFGIIPQEPVLFEGTVRSNIDPTGQYTDDEIWKSLDRCQLK 2373
Query: 451 ---------------QNGTQLSGGQKQRIAIARAILKNPKILLLDEATSALDAESEHIVQ 495
NG S GQ+Q + + R +LK ++L +DEAT+++D++++ ++Q
Sbjct: 2374 DTVASKPEKLDSLVVDNGDNWSVGQRQLLCLGRVMLKQSRLLFMDEATASVDSQTDAVIQ 2553
Query: 496 EALEKIILKRTTIVVAHRLTTIIHADTIAVVQQGKIVERGTHSEL 540
+ + + RT I +AHR+ T++ D + VV G+ E S L
Sbjct: 2554 KIIREDFAARTIISIAHRIPTVMDCDRVLVVDAGRAKEFDKPSNL 2688
Score = 108 bits (269), Expect = 1e-23
Identities = 82/288 (28%), Positives = 140/288 (48%), Gaps = 5/288 (1%)
Frame = +1
Query: 796 FPDTNKAIDSAASIFNILDSKPDIDSSSNDGVTQETVVGNI---ELQHVNFSYPTRPDIQ 852
FP + ++ A LD S+D V + + ++Q FS+ Q
Sbjct: 25 FPQSMISLSQALVSLGRLDRYMSSRELSDDSVERNEGCDGVIAVDVQDGTFSWDDEGLEQ 204
Query: 853 IFKDLTLSIPSAKTVALVGESGSGKSTVISLLERFYDPNSGRVLLDGVDIKTFRISWLRQ 912
K++ L + + A+VG GSGKS++++ + NSG+V + G + SW++
Sbjct: 205 DLKNINLKVNKGELTAIVGTVGSGKSSLLASILGEMHRNSGKVQVCGSTAYVAQTSWIQ- 381
Query: 913 QMGLVGQEPILFNESIRANIAYGKEDGATE-DEIIAAANAANAHNFISSLPDGYDTSVGE 971
N +I NI +G + +EII + + G T +GE
Sbjct: 382 ------------NGTIEENILFGLPMNRQKYNEII---RVCCLEKDLEMMEYGDQTEIGE 516
Query: 972 RGTQLSGGQKQRIAIARAMLKNPKILLLDEATSALDAES-ERIVQEALDRVSLNRTTVIV 1030
RG LSGGQKQRI +ARA+ ++ I LLD+ SA+DA + I +E + +T V+V
Sbjct: 517 RGINLSGGQKQRIQLARAVYQDCDIYLLDDVFSAVDAHTGTEIFKECVRGALKGKTIVLV 696
Query: 1031 AHRLTTIRGADTIAVIKNGMVAEKGRHDELMNNTHGVYASLVALHSTA 1078
H++ + D I V+++GM+ + GR+++L+++ + LVA H T+
Sbjct: 697 THQVDFLHNVDRIVVMRDGMIVQSGRYNDLLDSGLD-FGVLVAAHETS 837
Score = 77.8 bits (190), Expect = 2e-14
Identities = 56/211 (26%), Positives = 100/211 (46%), Gaps = 35/211 (16%)
Frame = +1
Query: 365 IELRDVHFRYPARPDVEIFAGFSLFVPSGTTTALVGQSGSGKSTVISLLERFYDPNAGEV 424
++++D F + + +L V G TA+VG GSGKS++++ + N+G+V
Sbjct: 154 VDVQDGTFSWDDEGLEQDLKNINLKVNKGELTAIVGTVGSGKSSLLASILGEMHRNSGKV 333
Query: 425 LIDGVNLKNLQLRWIRE----------------------------------QIGLVSQER 450
+ G Q WI+ + G ++
Sbjct: 334 QVCGSTAYVAQTSWIQNGTIEENILFGLPMNRQKYNEIIRVCCLEKDLEMMEYGDQTEIG 513
Query: 451 QNGTQLSGGQKQRIAIARAILKNPKILLLDEATSALDAES-EHIVQEALEKIILKRTTIV 509
+ G LSGGQKQRI +ARA+ ++ I LLD+ SA+DA + I +E + + +T ++
Sbjct: 514 ERGINLSGGQKQRIQLARAVYQDCDIYLLDDVFSAVDAHTGTEIFKECVRGALKGKTIVL 693
Query: 510 VAHRLTTIIHADTIAVVQQGKIVERGTHSEL 540
V H++ + + D I V++ G IV+ G +++L
Sbjct: 694 VTHQVDFLHNVDRIVVMRDGMIVQSGRYNDL 786
>TC87490 similar to PIR|T52080|T52080 multi resistance protein [imported] -
Arabidopsis thaliana, partial (19%)
Length = 927
Score = 132 bits (331), Expect = 9e-31
Identities = 76/248 (30%), Positives = 136/248 (54%)
Frame = +1
Query: 832 VVGNIELQHVNFSYPTRPDIQIFKDLTLSIPSAKTVALVGESGSGKSTVISLLERFYDPN 891
V G I+L + Y + + ++ + P K + +VG +GSGKST+I L R +P
Sbjct: 154 VNGTIQLIDLKVRYKENLPM-VLHGVSCTFPGGKKIGIVGRTGSGKSTLIQALFRLVEPA 330
Query: 892 SGRVLLDGVDIKTFRISWLRQQMGLVGQEPILFNESIRANIAYGKEDGATEDEIIAAANA 951
+G +L+D +DI + LR + ++ Q+P LF +IR N+ +E ++ EI A +
Sbjct: 331 AGSILIDNIDISGIGLHDLRSHLSIIPQDPTLFEGTIRGNLDPLEEH--SDKEIWEALDK 504
Query: 952 ANAHNFISSLPDGYDTSVGERGTQLSGGQKQRIAIARAMLKNPKILLLDEATSALDAESE 1011
+ I DT V + G S GQ+Q +A+ RA+LK KIL+LDEAT+++D+ ++
Sbjct: 505 SQLGEIIREKGQKLDTPVLQNGDNWSVGQRQLVALGRALLKQSKILVLDEATASVDSATD 684
Query: 1012 RIVQEALDRVSLNRTTVIVAHRLTTIRGADTIAVIKNGMVAEKGRHDELMNNTHGVYASL 1071
++Q+ + + T +AHR+ T+ +D + V+ +G++AE L+ + ++ L
Sbjct: 685 NLIQKVIREEFRDCTVCTIAHRIPTVIDSDLVLVLNDGLLAEFDTPLRLLEDKSSMFLKL 864
Query: 1072 VALHSTAS 1079
V +S+ S
Sbjct: 865 VTEYSSRS 888
Score = 108 bits (269), Expect = 1e-23
Identities = 66/239 (27%), Positives = 113/239 (46%), Gaps = 47/239 (19%)
Frame = +1
Query: 361 IKGDIELRDVHFRYPARPDVEIFAGFSLFVPSGTTTALVGQSGSGKSTVISLLERFYDPN 420
+ G I+L D+ RY + + G S P G +VG++GSGKST+I L R +P
Sbjct: 154 VNGTIQLIDLKVRYKENLPM-VLHGVSCTFPGGKKIGIVGRTGSGKSTLIQALFRLVEPA 330
Query: 421 AGEVLIDGVNLKNLQLRWIREQIGLVSQER------------------------------ 450
AG +LID +++ + L +R + ++ Q+
Sbjct: 331 AGSILIDNIDISGIGLHDLRSHLSIIPQDPTLFEGTIRGNLDPLEEHSDKEIWEALDKSQ 510
Query: 451 -----------------QNGTQLSGGQKQRIAIARAILKNPKILLLDEATSALDAESEHI 493
QNG S GQ+Q +A+ RA+LK KIL+LDEAT+++D+ ++++
Sbjct: 511 LGEIIREKGQKLDTPVLQNGDNWSVGQRQLVALGRALLKQSKILVLDEATASVDSATDNL 690
Query: 494 VQEALEKIILKRTTIVVAHRLTTIIHADTIAVVQQGKIVERGTHSELTMDPHGAYSQLI 552
+Q+ + + T +AHR+ T+I +D + V+ G + E T L D + +L+
Sbjct: 691 IQKVIREEFRDCTVCTIAHRIPTVIDSDLVLVLNDGLLAEFDTPLRLLEDKSSMFLKLV 867
>TC92097 similar to GP|22553016|emb|CAD44995. multidrug-resistance related
protein {Arabidopsis thaliana}, partial (25%)
Length = 1011
Score = 130 bits (327), Expect = 3e-30
Identities = 77/221 (34%), Positives = 123/221 (54%), Gaps = 1/221 (0%)
Frame = +2
Query: 834 GNIELQHVNFSY-PTRPDIQIFKDLTLSIPSAKTVALVGESGSGKSTVISLLERFYDPNS 892
G I+LQ + Y P P + K + + V +VG +GSGKST+IS L R +P+
Sbjct: 350 GKIDLQGLEIRYRPNSP--LVLKGIICTFKEGSRVGVVGRTGSGKSTLISALFRLVEPSR 523
Query: 893 GRVLLDGVDIKTFRISWLRQQMGLVGQEPILFNESIRANIAYGKEDGATEDEIIAAANAA 952
G +L+DG++I + + LR ++ ++ QEP LF SIR N+ ++DEI A
Sbjct: 524 GDILIDGINICSIGLKDLRTKLSIIPQEPTLFKGSIRTNL--DPLGLYSDDEIWKAVEKC 697
Query: 953 NAHNFISSLPDGYDTSVGERGTQLSGGQKQRIAIARAMLKNPKILLLDEATSALDAESER 1012
IS LP D+SV + G S GQ+Q + R +LK +IL+LDEAT+++D+ ++
Sbjct: 698 QLKETISKLPSLLDSSVSDEGGNWSLGQRQLFCLGRVLLKRNRILVLDEATASIDSATDA 877
Query: 1013 IVQEALDRVSLNRTTVIVAHRLTTIRGADTIAVIKNGMVAE 1053
I+Q + + T + VAHR+ T+ +D + V+ G + E
Sbjct: 878 ILQRIIRQEFEECTVITVAHRVPTVIDSDMVMVLSYGKLVE 1000
Score = 102 bits (255), Expect = 6e-22
Identities = 65/220 (29%), Positives = 110/220 (49%), Gaps = 48/220 (21%)
Frame = +2
Query: 362 KGDIELRDVHFRYPARPDVE-IFAGFSLFVPSGTTTALVGQSGSGKSTVISLLERFYDPN 420
KG I+L+ + RY RP+ + G G+ +VG++GSGKST+IS L R +P+
Sbjct: 347 KGKIDLQGLEIRY--RPNSPLVLKGIICTFKEGSRVGVVGRTGSGKSTLISALFRLVEPS 520
Query: 421 AGEVLIDGVNLKNLQLRWIREQIGLVSQE------------------------------- 449
G++LIDG+N+ ++ L+ +R ++ ++ QE
Sbjct: 521 RGDILIDGINICSIGLKDLRTKLSIIPQEPTLFKGSIRTNLDPLGLYSDDEIWKAVEKCQ 700
Query: 450 ----------------RQNGTQLSGGQKQRIAIARAILKNPKILLLDEATSALDAESEHI 493
G S GQ+Q + R +LK +IL+LDEAT+++D+ ++ I
Sbjct: 701 LKETISKLPSLLDSSVSDEGGNWSLGQRQLFCLGRVLLKRNRILVLDEATASIDSATDAI 880
Query: 494 VQEALEKIILKRTTIVVAHRLTTIIHADTIAVVQQGKIVE 533
+Q + + + T I VAHR+ T+I +D + V+ GK+VE
Sbjct: 881 LQRIIRQEFEECTVITVAHRVPTVIDSDMVMVLSYGKLVE 1000
>BQ751194 similar to GP|21622336|em probable multidrug resistance protein 2
{Neurospora crassa}, partial (16%)
Length = 808
Score = 130 bits (326), Expect = 4e-30
Identities = 74/128 (57%), Positives = 93/128 (71%), Gaps = 2/128 (1%)
Frame = -2
Query: 947 AAANAANAHNFISSLPDGYDTSVGERGTQLSGGQKQRIAIARAMLKNPKILLLDEATSAL 1006
+AA AN NFIS LP G DT VG RG+QLSGGQKQRIAIARA++K+P IL+LDEATSAL
Sbjct: 807 SAARKANC-NFISDLPAGLDTQVGARGSQLSGGQKQRIAIARALIKDPDILILDEATSAL 631
Query: 1007 DAESERIVQEALDRVSLNR-TTVIVAHRLTTIRGADTIAVIKN-GMVAEKGRHDELMNNT 1064
DAESE +V AL + R TT+ +AHRL+TI+ +D I V+ N G VAE G++ EL N
Sbjct: 630 DAESETLVNAALASLLKGRNTTISIAHRLSTIKRSDLIIVLSNDGKVAEIGKYSELSANK 451
Query: 1065 HGVYASLV 1072
++ L+
Sbjct: 450 ESAFSKLM 427
Score = 114 bits (285), Expect = 2e-25
Identities = 80/200 (40%), Positives = 111/200 (55%), Gaps = 3/200 (1%)
Frame = -2
Query: 444 GLVSQERQNGTQLSGGQKQRIAIARAILKNPKILLLDEATSALDAESEHIVQEALEKIIL 503
GL +Q G+QLSGGQKQRIAIARA++K+P IL+LDEATSALDAESE +V AL ++
Sbjct: 759 GLDTQVGARGSQLSGGQKQRIAIARALIKDPDILILDEATSALDAESETLVNAALASLLK 580
Query: 504 KR-TTIVVAHRLTTIIHADTIAVV-QQGKIVERGTHSELTMDPHGAYSQLIRLQEGEKEA 561
R TTI +AHRL+TI +D I V+ GK+ E G +SEL+ + A+S+L+ Q E
Sbjct: 579 GRNTTISIAHRLSTIKRSDLIIVLSNDGKVAEIGKYSELSANKESAFSKLMEWQMSGGEV 400
Query: 562 EGSRSSEVDKFGDNLNIDIHMAGSSTQRISFVRSISQTSSMSHRHSQLSGEIVDANIEQG 621
SR GD G T+ S+ + S + + ++G
Sbjct: 399 PESR-------GDR--------GHITEEEEIESSLEREDSEDTEARDAKKDATEEQ-QRG 268
Query: 622 QV-DNNEKPKMSMKNSIWRL 640
+V DN+ K + IWR+
Sbjct: 267 KVEDNSRK*SANHNGGIWRI 208
>BE943010 similar to GP|6634763|gb Similar to gb|AF008124 Arabidopsis thaliana
glutathione S-conjugate transporting ATPase (AtMRP1),
partial (15%)
Length = 629
Score = 127 bits (319), Expect = 2e-29
Identities = 68/211 (32%), Positives = 125/211 (59%)
Frame = +2
Query: 853 IFKDLTLSIPSAKTVALVGESGSGKSTVISLLERFYDPNSGRVLLDGVDIKTFRISWLRQ 912
+ L+ + S + + +VG +G+GKS++++ L R + SGR+++DG DI TF ++ LR+
Sbjct: 2 VLHGLSFVVSSMEKIGVVGRTGAGKSSMLNALFRIVELQSGRIIIDGCDISTFGLADLRR 181
Query: 913 QMGLVGQEPILFNESIRANIAYGKEDGATEDEIIAAANAANAHNFISSLPDGYDTSVGER 972
+ ++ Q P+LF+ ++R N+ E + +I A A+ + I G D V E
Sbjct: 182 VLTIIPQSPVLFSGTVRFNLDPFNE--YNDVDIWEALERAHMKDVIRRNQFGLDAQVSEG 355
Query: 973 GTQLSGGQKQRIAIARAMLKNPKILLLDEATSALDAESERIVQEALDRVSLNRTTVIVAH 1032
G S GQ+Q +++ARA+L+ K+L+LDEAT+++D ++ ++Q+ + + + T +I+AH
Sbjct: 356 GDNFSVGQRQLLSLARALLRRSKVLVLDEATASVDVRTDALIQKTIRQECNSCTMLIIAH 535
Query: 1033 RLTTIRGADTIAVIKNGMVAEKGRHDELMNN 1063
RL T+ + I ++ G V E EL+ N
Sbjct: 536 RLNTVVDCNRILLLDAGKVLEYNSPKELLQN 628
Score = 88.6 bits (218), Expect = 1e-17
Identities = 52/206 (25%), Positives = 99/206 (47%), Gaps = 47/206 (22%)
Frame = +2
Query: 382 IFAGFSLFVPSGTTTALVGQSGSGKSTVISLLERFYDPNAGEVLIDGVNLKNLQLRWIRE 441
+ G S V S +VG++G+GKS++++ L R + +G ++IDG ++ L +R
Sbjct: 2 VLHGLSFVVSSMEKIGVVGRTGAGKSSMLNALFRIVELQSGRIIIDGCDISTFGLADLRR 181
Query: 442 -----------------------------------------------QIGLVSQERQNGT 454
Q GL +Q + G
Sbjct: 182 VLTIIPQSPVLFSGTVRFNLDPFNEYNDVDIWEALERAHMKDVIRRNQFGLDAQVSEGGD 361
Query: 455 QLSGGQKQRIAIARAILKNPKILLLDEATSALDAESEHIVQEALEKIILKRTTIVVAHRL 514
S GQ+Q +++ARA+L+ K+L+LDEAT+++D ++ ++Q+ + + T +++AHRL
Sbjct: 362 NFSVGQRQLLSLARALLRRSKVLVLDEATASVDVRTDALIQKTIRQECNSCTMLIIAHRL 541
Query: 515 TTIIHADTIAVVQQGKIVERGTHSEL 540
T++ + I ++ GK++E + EL
Sbjct: 542 NTVVDCNRILLLDAGKVLEYNSPKEL 619
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.319 0.134 0.374
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 26,742,714
Number of Sequences: 36976
Number of extensions: 322155
Number of successful extensions: 2003
Number of sequences better than 10.0: 128
Number of HSP's better than 10.0 without gapping: 1746
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1959
length of query: 1079
length of database: 9,014,727
effective HSP length: 106
effective length of query: 973
effective length of database: 5,095,271
effective search space: 4957698683
effective search space used: 4957698683
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 64 (29.3 bits)
Medicago: description of AC140022.3


