

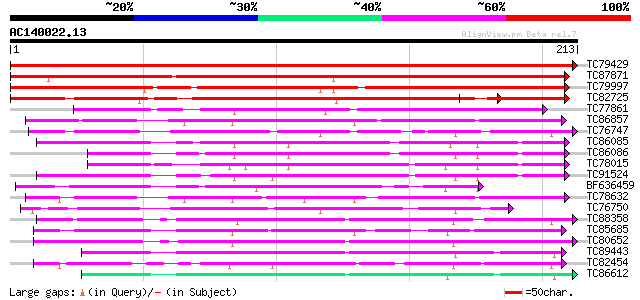
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140022.13 + phase: 0
(213 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC79429 similar to PIR|T03803|T03803 tumor-related protein clon... 444 e-125
TC87871 similar to PIR|T03803|T03803 tumor-related protein clon... 233 3e-62
TC79997 similar to PIR|T03803|T03803 tumor-related protein clon... 209 5e-55
TC82725 weakly similar to PIR|T07871|T07871 miraculin homolog r... 156 6e-39
TC77861 weakly similar to GP|13899081|gb|AAK48962.1 Unknown prot... 149 1e-36
TC86857 similar to GP|14161088|emb|CAB76907. alpha-fucosidase {C... 92 2e-19
TC76747 similar to GP|20269067|emb|CAD29731. protease inhibitor ... 89 1e-18
TC86085 86 8e-18
TC86086 similar to GP|15155442|gb|AAK86324.1 AGR_C_899p {Agrobac... 82 2e-16
TC78015 similar to GP|12721693|gb|AAK03411.1 RbsC {Pasteurella m... 80 6e-16
TC91524 similar to GP|15217238|gb|AAK92582.1 Hypothetical protei... 79 1e-15
BF636459 73 9e-14
TC78632 similar to GP|9367042|gb|AAF87095.1| trypsin inhibitor {... 72 2e-13
TC76750 similar to GP|20269067|emb|CAD29731. protease inhibitor ... 71 3e-13
TC88358 proteinase inhibitor 20 [Medicago truncatula] 67 5e-12
TC85685 similar to GP|14161088|emb|CAB76907. alpha-fucosidase {C... 63 1e-10
TC80652 similar to GP|14161088|emb|CAB76907. alpha-fucosidase {C... 61 4e-10
TC89443 similar to GP|7208767|emb|CAB76906.1 alpha-fucosidase {C... 59 1e-09
TC82454 weakly similar to GP|3551243|dbj|BAA32820.1 Water-Solubl... 58 2e-09
TC86612 weakly similar to GP|7208767|emb|CAB76906.1 alpha-fucosi... 57 4e-09
>TC79429 similar to PIR|T03803|T03803 tumor-related protein clone NF34 -
common tobacco, partial (42%)
Length = 807
Score = 444 bits (1142), Expect = e-125
Identities = 213/213 (100%), Positives = 213/213 (100%)
Frame = +2
Query: 1 MKSICILLAVLFALSTQPLLGEADASPEQVVDTEGKKVRAGVDYYIRPVPTTPCDGRGPC 60
MKSICILLAVLFALSTQPLLGEADASPEQVVDTEGKKVRAGVDYYIRPVPTTPCDGRGPC
Sbjct: 23 MKSICILLAVLFALSTQPLLGEADASPEQVVDTEGKKVRAGVDYYIRPVPTTPCDGRGPC 202
Query: 61 VVGSGFVLIARSPNETCPLNVVVVEGFRGQGVTFTPVNPKKGVIRVSTDLNIKTSLNTSC 120
VVGSGFVLIARSPNETCPLNVVVVEGFRGQGVTFTPVNPKKGVIRVSTDLNIKTSLNTSC
Sbjct: 203 VVGSGFVLIARSPNETCPLNVVVVEGFRGQGVTFTPVNPKKGVIRVSTDLNIKTSLNTSC 382
Query: 121 EESTIWTLDDFDSSTGQWFVTTGGVLGNPGKDTVDNWFKIEKYEDDYKFVFCPTVCNFCK 180
EESTIWTLDDFDSSTGQWFVTTGGVLGNPGKDTVDNWFKIEKYEDDYKFVFCPTVCNFCK
Sbjct: 383 EESTIWTLDDFDSSTGQWFVTTGGVLGNPGKDTVDNWFKIEKYEDDYKFVFCPTVCNFCK 562
Query: 181 VMCRNVGIFRDSNGNQRVALTDVPYKVRFQPSA 213
VMCRNVGIFRDSNGNQRVALTDVPYKVRFQPSA
Sbjct: 563 VMCRNVGIFRDSNGNQRVALTDVPYKVRFQPSA 661
>TC87871 similar to PIR|T03803|T03803 tumor-related protein clone NF34 -
common tobacco, partial (36%)
Length = 905
Score = 233 bits (595), Expect = 3e-62
Identities = 122/212 (57%), Positives = 149/212 (69%), Gaps = 2/212 (0%)
Frame = +3
Query: 1 MKSICILLAVLFA-LSTQPLLGEADASPEQVVDTEGKKVRAGVDYYIRPVPTTPCDGRGP 59
MK+ + LF LS+QPLLG A+AS EQVVDT GKK+RA +YYI PVP C G
Sbjct: 15 MKNTLLAFFFLFTFLSSQPLLGAAEASNEQVVDTLGKKLRADANYYIIPVPIYKCGPYGK 194
Query: 60 CVVGSGFVLIARSPNETCPLNVVVVEGFRGQGVTFTPVNPKKGVIRVSTDLNIKTSLNTS 119
C SG L S +TCPL+VVVV+ ++ +TF PVNPKKGVIRVSTDLNIK S +
Sbjct: 195 CR-SSGSSLALASIGKTCPLDVVVVDRYQALPLTFIPVNPKKGVIRVSTDLNIKFSSRAT 371
Query: 120 C-EESTIWTLDDFDSSTGQWFVTTGGVLGNPGKDTVDNWFKIEKYEDDYKFVFCPTVCNF 178
C S +W LD F+ S QWF+T GGV GNPG +T++NWFKIEKY D YK VFCP+V
Sbjct: 372 CLHHSMVWKLDRFNVSKRQWFITIGGVAGNPGWETINNWFKIEKYGDAYKLVFCPSVVQS 551
Query: 179 CKVMCRNVGIFRDSNGNQRVALTDVPYKVRFQ 210
K MC++VG+F D NGN+R+AL+DVP KV+FQ
Sbjct: 552 FKHMCKDVGVFVDENGNKRLALSDVPLKVKFQ 647
>TC79997 similar to PIR|T03803|T03803 tumor-related protein clone NF34 -
common tobacco, partial (48%)
Length = 801
Score = 209 bits (533), Expect = 5e-55
Identities = 113/215 (52%), Positives = 146/215 (67%), Gaps = 5/215 (2%)
Frame = +2
Query: 1 MKSICILLAVLFALSTQPLLGEADASPEQVVDTEGKKVRAGVDYYIRPV-PTTPCDGRGP 59
MK + +LFALS+QPLLG A+ASP+QV+DT GKK+RA +YYI P P T C G
Sbjct: 41 MKGTMLAFLLLFALSSQPLLGSAEASPDQVIDTSGKKLRADTNYYIIPAKPFTTC-GFVS 217
Query: 60 CVVGSGFVLIARSPNETCPLNVVVVEGFRGQGVTFTPVNPKKGVIRVSTDLNIKTS---L 116
C G L + E+CPL+VVVV+ +G + FTPVN KKGV+RVSTDLNIK S
Sbjct: 218 CFNSGGIAL--ETVGESCPLDVVVVKHNQGLPLRFTPVNNKKGVVRVSTDLNIKFSNDAY 391
Query: 117 NTSC-EESTIWTLDDFDSSTGQWFVTTGGVLGNPGKDTVDNWFKIEKYEDDYKFVFCPTV 175
++ C S +W +D F S + FVTT GVLGNPG +T+ NWF+IEKYED YK V+CP V
Sbjct: 392 DSRCPNHSLVWKIDPF--SKEETFVTTNGVLGNPGSNTIHNWFQIEKYEDAYKLVYCPNV 565
Query: 176 CNFCKVMCRNVGIFRDSNGNQRVALTDVPYKVRFQ 210
C CK +C+++GI+ R+ALT+VP+KV+FQ
Sbjct: 566 CPSCKHVCKDIGIYVYKYREMRLALTNVPFKVKFQ 670
>TC82725 weakly similar to PIR|T07871|T07871 miraculin homolog root-knot
nematode-induced - tomato, partial (51%)
Length = 818
Score = 156 bits (394), Expect = 6e-39
Identities = 86/187 (45%), Positives = 118/187 (62%), Gaps = 2/187 (1%)
Frame = +2
Query: 1 MKSICILLAVLFALSTQPLLGEADASPEQVVDTEGKKVRAGVDYYIR-PVPTTPCDGRGP 59
MKS + +L AL++QPLL +S E VVD GK +RA Y + +P T + R P
Sbjct: 59 MKSTFLAFLLLIALTSQPLLS---SSLEHVVDITGKNLRANAYYNVLLSMPYT--NSRSP 223
Query: 60 CVVGSGFVLIARSPNETCPLNVVVVEGFRGQGVTFTPVNPKKGVIRVSTDLNIKTSLNTS 119
+G ++ + + CPL+V+VV ++ + FTP+N KKGVIRVS+DLNI N+S
Sbjct: 224 EGLG-----LSNNIGQPCPLDVIVVSRYQSLPIRFTPLNLKKGVIRVSSDLNIMFRSNSS 388
Query: 120 CE-ESTIWTLDDFDSSTGQWFVTTGGVLGNPGKDTVDNWFKIEKYEDDYKFVFCPTVCNF 178
C +T+W LD FD+S G+ FVTT G +GNPG ++ NWFKIEKY + YK V+CP VC
Sbjct: 389 CPYHTTVWKLDRFDASKGKSFVTTDGFIGNPGPQSISNWFKIEKYVEGYKLVYCPIVCPS 568
Query: 179 CKVMCRN 185
CK C+N
Sbjct: 569 CKHECKN 589
Score = 42.0 bits (97), Expect = 2e-04
Identities = 19/41 (46%), Positives = 29/41 (70%)
Frame = +3
Query: 170 VFCPTVCNFCKVMCRNVGIFRDSNGNQRVALTDVPYKVRFQ 210
+F P V + + VG+F D NGN+R+AL+DVPY+V+F+
Sbjct: 555 LFVPLV----NMSAKMVGLFEDENGNKRLALSDVPYQVKFR 665
>TC77861 weakly similar to GP|13899081|gb|AAK48962.1 Unknown protein
{Arabidopsis thaliana}, partial (39%)
Length = 871
Score = 149 bits (375), Expect = 1e-36
Identities = 84/181 (46%), Positives = 106/181 (58%), Gaps = 3/181 (1%)
Frame = +3
Query: 25 ASPEQVVDTEGKKVRAGVDYYIRPVPTTPCDGRGPCVVGSGFVLIARSPNETCPLNVVV- 83
A+PE V+D GK++ GV YYI PV G G V G N+TCPL VV
Sbjct: 111 AAPEPVLDISGKQLTTGVKYYILPVIRGK--GGGLTVANHG------ENNQTCPLYVVQE 266
Query: 84 -VEGFRGQGVTFTPVNPKKGVIRVSTDLNIKTSLN-TSCEESTIWTLDDFDSSTGQWFVT 141
+E G+ VTFTP N K+GVI STDLNIK+ + T C ++ +W L TG WF+
Sbjct: 267 KLEVKNGEAVTFTPYNAKQGVILTSTDLNIKSFVTKTKCPQTQVWKL--LKELTGVWFLA 440
Query: 142 TGGVLGNPGKDTVDNWFKIEKYEDDYKFVFCPTVCNFCKVMCRNVGIFRDSNGNQRVALT 201
TGGV GNP TV NWFKIEK + DY FCP C+ +CR +G+F D GN+ +AL+
Sbjct: 441 TGGVEGNPSMATVGNWFKIEKADKDYVLSFCPAEACKCQTLCRELGLFVDDKGNKHLALS 620
Query: 202 D 202
D
Sbjct: 621 D 623
>TC86857 similar to GP|14161088|emb|CAB76907. alpha-fucosidase {Cicer
arietinum}, partial (41%)
Length = 860
Score = 91.7 bits (226), Expect = 2e-19
Identities = 70/208 (33%), Positives = 98/208 (46%), Gaps = 5/208 (2%)
Frame = +1
Query: 7 LLAVLFALSTQPLLGEADASPEQVVDTEGKKVRAGVDYYIRPVPTTPCDGRGPCVVGS-- 64
L +LFAL+ L A S EQV DT G + G +YI P + G+
Sbjct: 100 LSLILFALTICFSLAFAQVS-EQVFDTNGNPIFPGGTFYIMP-----------SIFGAAG 243
Query: 65 GFVLIARSPNETCPLNVV--VVEGFRGQGVTFTPVNPKKGVIRVSTDLNIKTSLNTSCEE 122
G + + ++ N CPL V+ E G V FT + +I +T L+I + C E
Sbjct: 244 GGLRLGKTKNSKCPLTVLQDYSEVVNGLPVKFTRLEAGHDIISTNTALDIAFTTKPDCAE 423
Query: 123 STIWTL-DDFDSSTGQWFVTTGGVLGNPGKDTVDNWFKIEKYEDDYKFVFCPTVCNFCKV 181
S+ W L DDF+ TG W V GG N GK ++ +FKI+K+ YK VFCP +
Sbjct: 424 SSKWVLVDDFNKLTGPW-VGIGGTEDNEGKHIINGFFKIQKHGFGYKLVFCPDI-TAPPG 597
Query: 182 MCRNVGIFRDSNGNQRVALTDVPYKVRF 209
C ++G D G V + PY+V F
Sbjct: 598 ACYDIGRHDDFTGRLLVLANNDPYEVVF 681
>TC76747 similar to GP|20269067|emb|CAD29731. protease inhibitor {Sesbania
rostrata}, partial (86%)
Length = 1189
Score = 89.4 bits (220), Expect = 1e-18
Identities = 72/215 (33%), Positives = 105/215 (48%), Gaps = 9/215 (4%)
Frame = +3
Query: 8 LAVLFALSTQPLLGEADASPEQVVDTEGKKVRAGVDYYIRPVPTTPCDGRGPCVVGSGFV 67
L++L +T+PL G A PE VVD +G ++ G YY+ P+ +G +
Sbjct: 129 LSLLVLFNTKPLQG---AEPEAVVDKQGNPLKPGEGYYVFPL-----------WADNGGI 266
Query: 68 LIARSPNETCPLNVVVVEGFRGQGVTFTPVNPKKGVIRVSTDLNIKTS-LNTSCEESTIW 126
+ + N+TCPL+V+ G V F+ I TDL I+ L + C E +W
Sbjct: 267 TLGHTRNKTCPLDVIRNPDAIGTPVYFSASG--LDYIPTLTDLTIEIPILGSPCNEPKVW 440
Query: 127 TLDDFDSSTGQWFVTTGGVLGNPGKDTVDNWFKIEKYEDD-----YKFVFCPTVCNFCKV 181
L +G WFV+TGG G D V FKIE+ + Y F FCP+V V
Sbjct: 441 RL--LKVGSGFWFVSTGGAAG----DLVSK-FKIERLAGEHAYEIYSFKFCPSVPG---V 590
Query: 182 MCRNVGIFRDSNGNQRVALTD---VPYKVRFQPSA 213
+C VG F D++G + +A+ D PY VRFQ ++
Sbjct: 591 LCAPVGTFVDTDGTKVMAVGDGIEEPYYVRFQKAS 695
>TC86085
Length = 884
Score = 86.3 bits (212), Expect = 8e-18
Identities = 67/207 (32%), Positives = 102/207 (48%), Gaps = 7/207 (3%)
Frame = +3
Query: 11 LFALSTQPLLGEADASPEQVVDTEGKKVRAGVDYYIRPVPTTPCDGRGPCVVGSGFVLIA 70
+F L+ LL + + V+DT G+ V +Y+IRP T G G L+
Sbjct: 81 IFILAHVWLLMATTSIAQFVIDTSGEPVEDDEEYFIRPAITGN---------GGGSTLV- 230
Query: 71 RSPNETCPLNVVV--VEGFRGQGVTFTPVNPKKGV--IRVSTDLNIKTSLNTSCEESTIW 126
+ N CPL+V + EG G V FTP P+ +R++ DL + +TSC +ST W
Sbjct: 231 -TGNGPCPLHVGLDNTEGTLGVAVKFTPFAPQHDDDDVRLNRDLRVTFLTSTSCGQSTDW 407
Query: 127 TLDDFDSSTGQWFVTTGGVLGNPGKDTVDNWFKIEKYE--DDYKFVFCPT-VCNFCKVMC 183
L + D+++G+ + TG + G + N+F+I + + Y +CPT C CKV C
Sbjct: 408 RLGEKDATSGRRLIVTG---RDNGAGSQGNFFRIVQTQTGGTYNIQWCPTEACPSCKVQC 578
Query: 184 RNVGIFRDSNGNQRVALTDVPYKVRFQ 210
VG+ R+ NG +AL V FQ
Sbjct: 579 GTVGVIRE-NGKNLLALDGDALPVVFQ 656
>TC86086 similar to GP|15155442|gb|AAK86324.1 AGR_C_899p {Agrobacterium
tumefaciens str. C58 (Cereon)}, partial (2%)
Length = 1329
Score = 82.0 bits (201), Expect = 2e-16
Identities = 63/190 (33%), Positives = 93/190 (48%), Gaps = 9/190 (4%)
Frame = -3
Query: 30 VVDTEGKKVRAGVDYYIRPVPTTPCDGRGPCVVGSGFVLIARSPNETCPLNVVV--VEGF 87
V+DT G+ V +Y+IRP T G G +L+ R N CPL+V + EG
Sbjct: 1186 VIDTSGEPVEDDEEYFIRPAITGN---------GGGSILVTR--NGPCPLHVGLGNSEGT 1040
Query: 88 RGQGVTFTPVNPKKGV----IRVSTDLNIKTSLNTSCEESTIWTLDDFDSSTGQWFVTTG 143
G V FTP P+ +R++ DL + T C +ST W L + D+++G+ + TG
Sbjct: 1039 LGLAVKFTPFAPRHDDDDDDVRLNRDLRVTFQGFTGCGQSTDWRLGEKDATSGRRLIVTG 860
Query: 144 GVLGNPGKDTVDNWFKIEKYEDD--YKFVFCPT-VCNFCKVMCRNVGIFRDSNGNQRVAL 200
+ G + N+F+I + + Y +CPT C CKV C VG+ R+ NG +AL
Sbjct: 859 ---RDNGAGSHGNFFRIVQTQTGGIYNIQWCPTEACPSCKVQCGTVGVIRE-NGKILLAL 692
Query: 201 TDVPYKVRFQ 210
V FQ
Sbjct: 691 DGGALPVVFQ 662
>TC78015 similar to GP|12721693|gb|AAK03411.1 RbsC {Pasteurella multocida},
partial (4%)
Length = 776
Score = 80.1 bits (196), Expect = 6e-16
Identities = 66/188 (35%), Positives = 93/188 (49%), Gaps = 7/188 (3%)
Frame = -3
Query: 30 VVDTEGKKVRAGVDYYIRPVPTTPCDGRGPCVVGSGFVLIARSPNETCPLNV-VVVEGF- 87
V+DT G+ V +Y+IRPV T GR V S NE+CPL+V + + G
Sbjct: 669 VLDTVGEPVEGDEEYFIRPVITNK-GGRSTMV----------SRNESCPLHVGLELTGLG 523
Query: 88 RGQGVTFTPVNPKKGV--IRVSTDLNIKTSLNTSCEESTIWTLDDFDSSTGQWFVTTGGV 145
RG V FTP P +RV+ DL I ++SC +ST W L + D+ +G+ + TG
Sbjct: 522 RGLVVKFTPFAPHHDFDDVRVNRDLRITFQASSSCVQSTEWRLGEKDTKSGRRLIITGTD 343
Query: 146 LGNPGKDTVDNWFKIEK--YEDDYKFVFCPT-VCNFCKVMCRNVGIFRDSNGNQRVALTD 202
G + N+F+I + E Y +CPT VC CK C V + + NG +AL
Sbjct: 342 SATNG--SYGNFFRIVETPLEGMYNIQWCPTEVCPSCKFECGTVDMLNE-NGKILLALDG 172
Query: 203 VPYKVRFQ 210
P + FQ
Sbjct: 171 GPLPLVFQ 148
>TC91524 similar to GP|15217238|gb|AAK92582.1 Hypothetical protein {Oryza
sativa} [Oryza sativa (japonica cultivar-group)],
partial (5%)
Length = 824
Score = 79.0 bits (193), Expect = 1e-15
Identities = 65/206 (31%), Positives = 98/206 (47%), Gaps = 6/206 (2%)
Frame = +3
Query: 11 LFALSTQPLLGEADASPEQVVDTEGKKVRAGVDYYIRPVPTTPCDGRGPCVVGSGFVLIA 70
+F L+ LL + + V+DT G+ V DY+IRP T G L+
Sbjct: 45 IFILANVWLLVVTTSIAQFVIDTSGEPVENDEDYFIRPAITGN---------GGSLTLVT 197
Query: 71 RSPNETCPLNVVV-VEGFRGQGVTFTPV--NPKKGVIRVSTDLNIKTSLNTSCEESTIWT 127
R+ +CP NV + + +G V +P N ++ +R+ DL + TSC +ST W
Sbjct: 198 RN---SCPFNVGLDPDAPQGFAVLLSPFVSNREEDEVRLGRDLRVIFQAGTSCGQSTEWR 368
Query: 128 LDDFDSSTGQWFVTTGGVLGNPGKDTVDNWFKIEKYEDD--YKFVFCPT-VCNFCKVMCR 184
L + D++TG+ F+ TG G + N+F+I + + +CPT VC CK C
Sbjct: 369 LGERDATTGRRFIITGRDDSTVG--SYGNFFRIVQTPSRGIFNIQWCPTEVCPSCKFECG 542
Query: 185 NVGIFRDSNGNQRVALTDVPYKVRFQ 210
VGI R+ NG +AL V FQ
Sbjct: 543 TVGIVRE-NGKILLALDGSALPVAFQ 617
>BF636459
Length = 519
Score = 72.8 bits (177), Expect = 9e-14
Identities = 57/181 (31%), Positives = 89/181 (48%), Gaps = 5/181 (2%)
Frame = +1
Query: 3 SICILLAVLFALSTQPLLGEADASPEQVVDTEGKKVRAGVDYYIRPVPTTPCDGRGPCVV 62
S+ ++L + ++ T A + E+++DT+G + G +YYI+P T
Sbjct: 52 SLSLMLCLFMSIKTL-----AQSENEKILDTKGHPLERGKEYYIKPAITDS--------- 189
Query: 63 GSGFVLIARSPNETCPLNVVVVEGFRGQG--VTFTPVNPKKGVIRVSTDLNIKTSLNTSC 120
G F LI R N +CPL V G+G V FTP + VI+ S D +K S ++ C
Sbjct: 190 GGRFTLIDR--NGSCPLYVGQENTDLGKGLPVIFTPFAKEDKVIKDSRDFKVKFSASSIC 363
Query: 121 EESTIWTLDDFDSSTGQWFVTTGGVLGNPGKDTVDNWFKIEK--YEDDYKFVFCPT-VCN 177
+ST W L D D+ +G+ + + G+ G ++F+I K +E Y FCPT C
Sbjct: 364 VQSTEWKLGDRDTKSGRRVI----IAGSDG-----SYFRIVKAEFEGVYNIRFCPTDTCX 516
Query: 178 F 178
F
Sbjct: 517 F 519
>TC78632 similar to GP|9367042|gb|AAF87095.1| trypsin inhibitor {Glycine
max}, partial (44%)
Length = 803
Score = 72.0 bits (175), Expect = 2e-13
Identities = 67/214 (31%), Positives = 100/214 (46%), Gaps = 10/214 (4%)
Frame = +2
Query: 7 LLAVLFALSTQ-PLLGEADASPEQVVDTEGKKVRAGVDYYIRPVPTTPCDGRGPCVVGS- 64
L +LFAL+T PL A EQV D+ G + +Y++P + G+
Sbjct: 32 LSLLLFALATYFPL-----AFTEQVRDSNGNPIFFSSRFYVKP-----------SIFGAA 163
Query: 65 -GFVLIARSPNETCPLNVV--VVEGFRGQGVTFTPVNPKKGVIRVSTD---LNIKTSLNT 118
G V + + N +CPL V+ E G V F+ + + + +STD ++I
Sbjct: 164 GGGVKLGETGNSSCPLTVLQDYSEVVNGLPVKFS-TDAEIFIDLISTDTSRVDIVFPEKP 340
Query: 119 SCEESTIWTL--DDFDSSTGQWFVTTGGVLGNPGKDTVDNWFKIEKYEDDYKFVFCPTVC 176
C ES+ W L DDF +V GG+ GK +D FKI K+ YK VFCPT
Sbjct: 341 ECAESSKWLLIEDDFPRP----WVGIGGIEDYIGKHIIDGKFKIVKHGFGYKLVFCPTF- 505
Query: 177 NFCKVMCRNVGIFRDSNGNQRVALTDVPYKVRFQ 210
+C ++G + D NG + + D PY+V F+
Sbjct: 506 TAPPGLCHDIGRYDDKNGRRLILTEDDPYEVVFE 607
>TC76750 similar to GP|20269067|emb|CAD29731. protease inhibitor {Sesbania
rostrata}, partial (77%)
Length = 651
Score = 71.2 bits (173), Expect = 3e-13
Identities = 58/192 (30%), Positives = 91/192 (47%), Gaps = 7/192 (3%)
Frame = +3
Query: 5 CIL-LAVLFALSTQPLLGEADASPEQVVDTEGKKVRAGVDYYIRPVPTTPCDGRGPCVVG 63
C L L+ LF +T+ L G+ PE VVD +G ++ G YY+ P+
Sbjct: 18 CFLSLSALF--NTKALQGD---KPEAVVDKQGNPLKPGEGYYVFPL-----------WAD 149
Query: 64 SGFVLIARSPNETCPLNVVVVEGFRGQGVTFTPVNPKKGVIRVSTDLNIKTS-LNTSCEE 122
+G + + ++ N+TCPL+V+ G V F + I TDL ++ L + C E
Sbjct: 150 NGGITLGQTRNKTCPLDVIRNPEAIGSPVYFYEY--EHDYIPTLTDLTVEIPILGSPCSE 323
Query: 123 STIWTLDDFDSSTGQWFVTTGGVLGNPGKDTVDNWFKIEKYEDD-----YKFVFCPTVCN 177
+W + + WFV+TGG GN + + FKIE+ E + Y F++CP+V
Sbjct: 324 RKVWKISKEGTRARFWFVSTGGFPGN-----LFSQFKIERLEGEHAYEIYSFLYCPSVPG 488
Query: 178 FCKVMCRNVGIF 189
+C VG F
Sbjct: 489 ---TLCAPVGTF 515
>TC88358 proteinase inhibitor 20 [Medicago truncatula]
Length = 806
Score = 67.0 bits (162), Expect = 5e-12
Identities = 64/208 (30%), Positives = 91/208 (42%), Gaps = 5/208 (2%)
Frame = +2
Query: 11 LFALSTQPLLGEADASPEQVVDTEGKKVRAGVDYYIRPVPTTPCDGRGPCVVGSGFVLIA 70
+F T P L ++ EQV+D G + G YYI P RGP G G V +
Sbjct: 74 IFVFITNPSLATSN-DVEQVLDINGNPIFPGGQYYILPAL------RGP---GGGGVRLG 223
Query: 71 RSPNETCPLNVVVV--EGFRGQGVTFTPVNPKKGVIRVSTDLNIKTSLNTSCEESTIWTL 128
R+ + CP+ V+ E G V FT G+I T L I+ + SC ST W L
Sbjct: 224 RTGDLKCPVTVLQDRREVKNGLPVKFTIPGISPGIIFTGTPLEIEYTKKPSCAASTKW-L 400
Query: 129 DDFDSSTGQWFVTTGGVLGNPGKDTVDNWFKIEKYED--DYKFVFCPTVCNFCKVMCRNV 186
D+ G+ + GG PG T+ F I+K+ Y FC T C ++
Sbjct: 401 IFVDNVIGKACIGIGGPENYPGVQTLKGKFNIQKHASGFGYNLGFCVT----GSPTCLDI 568
Query: 187 GIFRDSNGNQRVALTD-VPYKVRFQPSA 213
G F + +R+ LT+ Y+V F +A
Sbjct: 569 GRFDNDEAGRRLNLTEHEVYQVVFVDAA 652
>TC85685 similar to GP|14161088|emb|CAB76907. alpha-fucosidase {Cicer
arietinum}, partial (42%)
Length = 870
Score = 62.8 bits (151), Expect = 1e-10
Identities = 62/207 (29%), Positives = 93/207 (43%), Gaps = 7/207 (3%)
Frame = +1
Query: 10 VLFALSTQPLLGEADASPEQVVDTEGKKVRAGVDYYIRPVPTTPCDGRGPCVVGSGFVLI 69
+LF+ + PL A E V D G V G YYI P+ + G G + +
Sbjct: 85 LLFSFTYFPL-----AFTETVEDINGNPVFPGGKYYIAPLISKG---------GGGGLKL 222
Query: 70 ARSPNETCPLNVV--VVEGFRGQGVTFTPVNPKKGVIRVSTDLNIKTSLNTSCEESTIWT 127
++ + CP+ V+ E RG V FT + K+GVI + +++I+ C ES W
Sbjct: 223 GKTGDSECPVTVLQDFSEVVRGLPVRFTII-VKRGVIFTTDEVDIEFVKKPKCAESAKWV 399
Query: 128 L--DDFDSSTGQWFVTTGGVLGNPGKDTVDNWFKIEKY---EDDYKFVFCPTVCNFCKVM 182
L DDF +S W + GK FKIE YK V+CP + +
Sbjct: 400 LAHDDFPTS---WVGIGDNIDAFQGK------FKIETLGSGSGAYKLVYCP-LFSAPPGA 549
Query: 183 CRNVGIFRDSNGNQRVALTDVPYKVRF 209
C ++G +RD NG + V + P++V F
Sbjct: 550 CSDIGRYRDENGWRLVPTENDPFRVVF 630
>TC80652 similar to GP|14161088|emb|CAB76907. alpha-fucosidase {Cicer
arietinum}, partial (81%)
Length = 788
Score = 60.8 bits (146), Expect = 4e-10
Identities = 59/208 (28%), Positives = 88/208 (41%), Gaps = 4/208 (1%)
Frame = +2
Query: 10 VLFALSTQPLLGEADASPEQVVDTEGKKVRAGVDYYIRPVPTTPCDGRGPCVVGSGFVLI 69
+LF +T L ++ + EQV+D G + G YYI P RGP G + +
Sbjct: 32 LLFVFTTNLSLAFSNDAVEQVLDINGNPIFPGGKYYILPAI------RGPL---GGGLRL 184
Query: 70 ARSPNETCPLNVV--VVEGFRGQGVTFTPVNPKKGVIRVSTDLNIKTSLNTSCEESTIWT 127
+S N C + VV E G V F+ G+I T ++I+ + +C ES+ W
Sbjct: 185 GKSSNSDCEVTVVQDYNEVINGVPVKFSIPEISPGIIFTGTPIDIEFTKKPNCVESSKW- 361
Query: 128 LDDFDSSTGQWFVTTGGVLGNPGKDTVDNWFKIEKYED--DYKFVFCPTVCNFCKVMCRN 185
L DS + V GG PG T+ F IEK+E Y+ +C C + R
Sbjct: 362 LIFVDSVIQKACVGIGGPENYPGFRTLSGTFNIEKHESGFGYRLGYCVKDSPTCLDIGRA 541
Query: 186 VGIFRDSNGNQRVALTDVPYKVRFQPSA 213
D G++ V + V F +A
Sbjct: 542 HEEVEDEGGSRLHLTHQVAFAVVFVDAA 625
>TC89443 similar to GP|7208767|emb|CAB76906.1 alpha-fucosidase {Cicer
arietinum}, partial (78%)
Length = 735
Score = 59.3 bits (142), Expect = 1e-09
Identities = 54/185 (29%), Positives = 75/185 (40%), Gaps = 3/185 (1%)
Frame = +3
Query: 28 EQVVDTEGKKVRAGVDYYIRPVPTTPCDGRGPCVVGSGFVLIARSPNETCPLNVVVVEGF 87
+QV+D G + G YYI P P SG + + + + CP+ V+
Sbjct: 108 KQVLDIHGTPLIPGSQYYIFPASENP---------NSGGLTLNKVGDLECPVTVLQNNAM 260
Query: 88 RGQGVTFTPVNPKKGVIRVSTDLNIKTSLNTSCEESTIWTLDDFDSSTGQWFVTTGGVLG 147
G V FT G I TDL I+ + C ES+ W L D +T V GG
Sbjct: 261 IGLPVKFTVPENNTGNILTGTDLEIEFTKKPDCAESSKW-LMFLDHNTQLSCVGIGGATN 437
Query: 148 NPGKDTVDNWFKIEKYEDD--YKFVFCPTVCNFCKVMCRNVGIFRDSNGNQRVALTDV-P 204
G +T+ F I K+ Y+ FC V C + + +F G R+ LT V
Sbjct: 438 YHGIETISGKFLIVKHGSGHVYRLGFCLDVTGDCGYI--GLQMFNSEEGGSRLFLTAVDA 611
Query: 205 YKVRF 209
Y V F
Sbjct: 612 YSVVF 626
>TC82454 weakly similar to GP|3551243|dbj|BAA32820.1 Water-Soluble
Chlorophyll Protein {Brassica oleracea}, partial (17%)
Length = 940
Score = 58.2 bits (139), Expect = 2e-09
Identities = 64/208 (30%), Positives = 86/208 (40%), Gaps = 8/208 (3%)
Frame = +2
Query: 10 VLFALSTQ-PLLGEADASPEQVVDTEGKKVRAGVDYYIRPVPTTPCDGRGPCVVGSGFVL 68
+LFAL+T PL EQV D G + Y+I P DG G G L
Sbjct: 71 LLFALTTYFPL--PFTQGVEQVKDKNGNPILVSKKYFI-----WPADGSG-----GGLRL 214
Query: 69 IARSPNETCPLNV--VVVEGFRGQGVTFTPV-NPKKGVIRVSTDLNIKTSLNTSCEESTI 125
+ E CPL V E + + F P N + T L+I T C ES+
Sbjct: 215 ---NETEQCPLVVQQAFSEDVKSLPLKFIPTENINDFIFTGYTSLDIVFEKKTKCAESSK 385
Query: 126 WTLDDFDSSTGQWFVTTGGVLGNPGKDTVDNWFKIEKYED--DYKFVFCPTVCNFCKVMC 183
W + W GGV GK +D FKIE YK VFCPT+ + C
Sbjct: 386 WVVVK-GGFMEPWIGIGGGV---NGKSVIDGLFKIETIRSFRGYKLVFCPTISD-PTGQC 550
Query: 184 RNVGIFRDSNGNQRVALTD--VPYKVRF 209
N+G F D+ R+ +++ P++V F
Sbjct: 551 NNIGRFFDNENGLRLIMSENFKPFEVVF 634
>TC86612 weakly similar to GP|7208767|emb|CAB76906.1 alpha-fucosidase {Cicer
arietinum}, partial (81%)
Length = 808
Score = 57.4 bits (137), Expect = 4e-09
Identities = 53/189 (28%), Positives = 73/189 (38%), Gaps = 3/189 (1%)
Frame = +3
Query: 28 EQVVDTEGKKVRAGVDYYIRPVPTTPCDGRGPCVVGSGFVLIARSPNETCPLNVVVVEGF 87
+ V D G + G YYI P P SG + + + + CPL V+
Sbjct: 120 QPVFDKHGNPLTPGNQYYILPASDNP---------SSGGLTLDKVGDSVCPLTVLQNNAV 272
Query: 88 RGQGVTFTPVNPKKGVIRVSTDLNIKTSLNTSCEESTIWTLDDFDSSTGQWFVTTGGVLG 147
G V FT + I TDL I+ C ES+ W L D T FV GG
Sbjct: 273 TGLPVKFTILENSTSNIVTGTDLEIEFINKPDCAESSKW-LMVVDHVTQLSFVGIGGPAN 449
Query: 148 NPGKDTVDNWFKIEKY--EDDYKFVFCPTVCNFCKVMCRNVGIFRDSNGNQRVALTDV-P 204
PG + + F I K+ + Y+ FC C + + F G R+ LT +
Sbjct: 450 YPGVELISGKFLILKHGSGNAYRVGFCLDTTGDCAYL--GLQEFNSGEGGSRLILTAINA 623
Query: 205 YKVRFQPSA 213
Y V F +A
Sbjct: 624 YSVVFVDAA 650
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.320 0.139 0.435
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,494,046
Number of Sequences: 36976
Number of extensions: 109427
Number of successful extensions: 495
Number of sequences better than 10.0: 49
Number of HSP's better than 10.0 without gapping: 471
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 471
length of query: 213
length of database: 9,014,727
effective HSP length: 92
effective length of query: 121
effective length of database: 5,612,935
effective search space: 679165135
effective search space used: 679165135
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 56 (26.2 bits)
Medicago: description of AC140022.13


