

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC139854.8 - phase: 0
(304 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
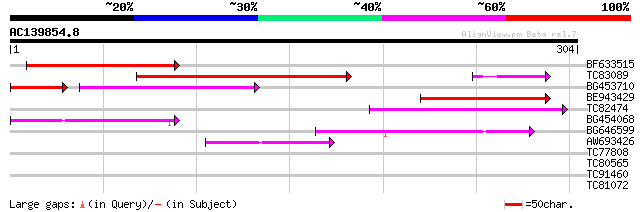
Score E
Sequences producing significant alignments: (bits) Value
BF633515 151 3e-37
TC83089 146 1e-35
BG453710 weakly similar to GP|21537057|gb| unknown {Arabidopsis ... 74 8e-22
BE943429 85 3e-17
TC82474 70 1e-12
BG454068 weakly similar to GP|22202804|dbj hypothetical protein~... 68 4e-12
BG646599 67 7e-12
AW693426 homologue to GP|23307580|dbj similar to serine/threonin... 44 1e-04
TC77808 similar to PIR|T03843|T03843 prohibitin - common tobacco... 30 1.5
TC80565 weakly similar to GP|14335072|gb|AAK59800.1 AT5g20050/F2... 28 5.7
TC91460 similar to GP|19698839|gb|AAL91155.1 cytochrome P450 {Ar... 27 7.5
TC81072 similar to PIR|T05401|T05401 hypothetical protein F10M6.... 27 7.5
>BF633515
Length = 249
Score = 151 bits (381), Expect = 3e-37
Identities = 71/82 (86%), Positives = 75/82 (90%)
Frame = +2
Query: 10 LDDVACLTHLPIEGRMLDHGKKMPKHEGAAQLMTYLGVSQHEAEKICNQKYGGYISYPRL 69
LDDVACLTHLPIEGRML HGKKMPKHEGAA LM +LGVSQ EAEKIC Q+YGGYISYPRL
Sbjct: 2 LDDVACLTHLPIEGRMLSHGKKMPKHEGAALLMRHLGVSQQEAEKICGQEYGGYISYPRL 181
Query: 70 RDFYTSYLGRANVLAGTEDPEE 91
RDFYT+YLGRAN+LA TEDP E
Sbjct: 182RDFYTTYLGRANLLASTEDPVE 247
>TC83089
Length = 887
Score = 146 bits (368), Expect = 1e-35
Identities = 72/116 (62%), Positives = 88/116 (75%), Gaps = 1/116 (0%)
Frame = +3
Query: 69 LRDFYTSYLGRANVLAGTEDPEELEELARVRTYCVRCYLLYLVGCLLFGDRSNKRIELIY 128
LR++Y YL A L+ ++ + +EL ++RT CV+ YLLYLVGCLLFGD+SNKRIEL+Y
Sbjct: 264 LREYYEGYLDSATRLSDPQNLGDPQELEKIRTTCVKWYLLYLVGCLLFGDKSNKRIELVY 443
Query: 129 LTTMADGYAGMRNYSWGGMTLAYLYGELADACRPGHRALGGSVTLLTT-WFLAHFP 183
LTTM DGYAGMRN+SWGGMTLAYLY L++ PG AL SVTL+T WFL H P
Sbjct: 444 LTTMEDGYAGMRNHSWGGMTLAYLYHCLSEVSLPGGGALERSVTLVTAGWFLCHLP 611
Score = 41.6 bits (96), Expect = 4e-04
Identities = 20/42 (47%), Positives = 25/42 (58%)
Frame = +3
Query: 249 SGWIMCGVRRVYRHLPKRVLRQYGYVQTIPRHPTDVRDLPTA 290
+GW +C HLP+R LR YGYVQT+P PT + L A
Sbjct: 585 AGWFLC-------HLPERGLR*YGYVQTVPIPPTTIEPLEPA 689
>BG453710 weakly similar to GP|21537057|gb| unknown {Arabidopsis thaliana},
partial (6%)
Length = 651
Score = 74.3 bits (181), Expect(2) = 8e-22
Identities = 45/98 (45%), Positives = 57/98 (57%), Gaps = 1/98 (1%)
Frame = -3
Query: 38 AAQLMT-YLGVSQHEAEKICNQKYGGYISYPRLRDFYTSYLGRANVLAGTEDPEELEELA 96
A LMT LG+S A + G+ISYP L+ Y +L A L EEL+E A
Sbjct: 307 AIDLMTRLLGMSDANAMAEIRTESAGHISYPTLKRVYEDHLIEARRLEDPLTREELQERA 128
Query: 97 RVRTYCVRCYLLYLVGCLLFGDRSNKRIELIYLTTMAD 134
R R +CVR LLYLV C+LF ++N+ I+LIYL MAD
Sbjct: 127 RRRQWCVRSLLLYLVDCVLFTYKTNRHIDLIYLDCMAD 14
Score = 46.6 bits (109), Expect(2) = 8e-22
Identities = 20/31 (64%), Positives = 25/31 (80%)
Frame = -1
Query: 1 MPMGEMTVTLDDVACLTHLPIEGRMLDHGKK 31
+PMGEMT+TLDDV L H+PI G MLDH ++
Sbjct: 414 LPMGEMTITLDDVYNLLHIPIHGCMLDHDEQ 322
>BE943429
Length = 384
Score = 85.1 bits (209), Expect = 3e-17
Identities = 36/70 (51%), Positives = 51/70 (72%)
Frame = +2
Query: 221 DRIQLDDVCYKPYKEHREIQDFEEVFWYSGWIMCGVRRVYRHLPKRVLRQYGYVQTIPRH 280
D ++ V ++PY+ R++ F++V WYSGWIM G +++ RHLP+RVLRQYGYVQT+PR
Sbjct: 2 DSMEHCHVIWRPYEHRRDVTPFQDVCWYSGWIMAGKQKMVRHLPERVLRQYGYVQTVPRP 181
Query: 281 PTDVRDLPTA 290
PT + L A
Sbjct: 182 PTTIVPLAPA 211
>TC82474
Length = 992
Score = 70.1 bits (170), Expect = 1e-12
Identities = 34/106 (32%), Positives = 55/106 (51%)
Frame = -2
Query: 194 YLENYPVAARWKLPKGHGEGITYWSLLDRIQLDDVCYKPYKEHREIQDFEEVFWYSGWIM 253
Y E+ P A R+ L GH Y + LDR+++DDV + Y H + F+++ + WI
Sbjct: 883 YTEDRPHATRFSLAMGHKSPNNYMTFLDRVEVDDVSFNQYDFHHQTCPFDDIS*FFRWIE 704
Query: 254 CGVRRVYRHLPKRVLRQYGYVQTIPRHPTDVRDLPTAFHCADVRRL 299
C + Y HL + V++ YG++Q+ RHP + D+ RL
Sbjct: 703 CDRKMNYHHLLELVMK*YGHIQSTLRHPLSRTRAKCLSNILDITRL 566
>BG454068 weakly similar to GP|22202804|dbj hypothetical protein~predicted by
GeneMark.hmm etc. {Oryza sativa (japonica
cultivar-group)}, partial (5%)
Length = 673
Score = 68.2 bits (165), Expect = 4e-12
Identities = 36/94 (38%), Positives = 52/94 (55%), Gaps = 3/94 (3%)
Frame = +3
Query: 1 MPMGEMTVTLDDVACLTHLPIEGRMLDHGKKMPKHEGAAQLMTYLGVSQHEAEKICNQKY 60
+P+GEMT+ LDD++CL H+P+ G +L H + + H+G L+ YLG+ E +
Sbjct: 387 LPVGEMTIALDDISCLLHIPVGGNLLFH-ESLSIHQGTEYLVNYLGLEFEEIAAETKRLK 563
Query: 61 GGYISYPRLRDFYTSYLGRANVLA---GTEDPEE 91
+I+Y L YTSYL A A G ED E
Sbjct: 564 SAHITYDTLLSIYTSYLTEAKSYANQPGEEDSME 665
>BG646599
Length = 711
Score = 67.4 bits (163), Expect = 7e-12
Identities = 40/121 (33%), Positives = 64/121 (52%), Gaps = 4/121 (3%)
Frame = +3
Query: 165 RALGGSVTLLTTWFLAHFPGFFSVDLNTDYLENYPV----AARWKLPKGHGEGITYWSLL 220
+ LGG TLL W +FP + N + + A RWK P+G+ + ++
Sbjct: 33 KQLGGYATLLQCWIYEYFPTLGNRVENRVACDEPGMGVARAMRWKYPRGNLKVDQIRRMI 212
Query: 221 DRIQLDDVCYKPYKEHREIQDFEEVFWYSGWIMCGVRRVYRHLPKRVLRQYGYVQTIPRH 280
D + +DV + P+ HR++ F+++ SG+I V +LP+R LRQ+GY+Q IPR
Sbjct: 213 DDLTPNDVIWCPFDSHRQVIPFDDICLSSGYIRW-CSNVVPYLPERCLRQFGYIQYIPRP 389
Query: 281 P 281
P
Sbjct: 390 P 392
>AW693426 homologue to GP|23307580|dbj similar to serine/threonine
phosphatase PP7 {Oryza sativa (japonica
cultivar-group)}, partial (2%)
Length = 209
Score = 43.5 bits (101), Expect = 1e-04
Identities = 25/69 (36%), Positives = 36/69 (51%)
Frame = +1
Query: 106 YLLYLVGCLLFGDRSNKRIELIYLTTMADGYAGMRNYSWGGMTLAYLYGELADACRPGHR 165
+LL LVGC LF D+S + + YL D Y+WG LA+LY L A +
Sbjct: 1 FLLCLVGCTLFSDKSTFVVNVAYLEFFRD-LDSCGGYAWGVAALAHLYDNLRYASFHHMK 177
Query: 166 ALGGSVTLL 174
++ G +TL+
Sbjct: 178 SISGYLTLI 204
>TC77808 similar to PIR|T03843|T03843 prohibitin - common tobacco, partial
(96%)
Length = 1202
Score = 29.6 bits (65), Expect = 1.5
Identities = 15/37 (40%), Positives = 19/37 (50%), Gaps = 1/37 (2%)
Frame = -3
Query: 226 DDVCYKPYKEHREIQDFEEVFWYSGWI-MCGVRRVYR 261
+DV Y+ +EH E E WY W+ MC V YR
Sbjct: 393 EDVQYQDEREHEE*DSPSEDLWYRRWMRMCEVECGYR 283
>TC80565 weakly similar to GP|14335072|gb|AAK59800.1 AT5g20050/F28I16_200
{Arabidopsis thaliana}, partial (81%)
Length = 1581
Score = 27.7 bits (60), Expect = 5.7
Identities = 16/42 (38%), Positives = 22/42 (52%), Gaps = 3/42 (7%)
Frame = +2
Query: 81 NVLAGTEDPEELEELARVRTYC---VRCYLLYLVGCLLFGDR 119
++LA EDP +L LAR+ T V C Y +G +F R
Sbjct: 1325 DLLADDEDPTDLNNLARLLTPVSSNVECTSTYSLGSTIFSGR 1450
>TC91460 similar to GP|19698839|gb|AAL91155.1 cytochrome P450 {Arabidopsis
thaliana}, partial (59%)
Length = 976
Score = 27.3 bits (59), Expect = 7.5
Identities = 19/53 (35%), Positives = 25/53 (46%), Gaps = 12/53 (22%)
Frame = +2
Query: 165 RALGGSVTLLTTW----FLAHFPGFFSVDLNTDYLE--------NYPVAARWK 205
RA G S T T+ FLAH GF++V + LE NYP +W+
Sbjct: 161 RATGVSATYQTSIMPFPFLAHKQGFYTVTCHPKNLEHILRTRFDNYPKGPKWQ 319
>TC81072 similar to PIR|T05401|T05401 hypothetical protein F10M6.90 -
Arabidopsis thaliana, partial (13%)
Length = 745
Score = 27.3 bits (59), Expect = 7.5
Identities = 10/39 (25%), Positives = 24/39 (60%), Gaps = 3/39 (7%)
Frame = +2
Query: 80 ANVLAGTEDPEELEELARV---RTYCVRCYLLYLVGCLL 115
A+++ ++ P + R+ +T+C+RC L+Y++ C +
Sbjct: 302 ASIIGSSQKPNR*QTFQRLCYDQTWCLRCNLVYVLRCFV 418
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.325 0.142 0.463
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,473,258
Number of Sequences: 36976
Number of extensions: 181635
Number of successful extensions: 958
Number of sequences better than 10.0: 24
Number of HSP's better than 10.0 without gapping: 952
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 957
length of query: 304
length of database: 9,014,727
effective HSP length: 96
effective length of query: 208
effective length of database: 5,465,031
effective search space: 1136726448
effective search space used: 1136726448
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.5 bits)
S2: 58 (26.9 bits)
Medicago: description of AC139854.8


