

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC139841.8 + phase: 0
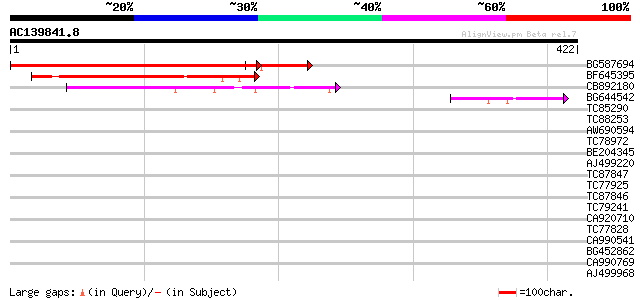
(422 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BG587694 similar to GP|20260480|gb unknown protein {Arabidopsis ... 385 e-107
BF645395 similar to GP|15808946|gb| auxin-regulated protein {Lyc... 147 1e-35
CB892180 similar to PIR|D84681|D84 hypothetical protein At2g2815... 144 5e-35
BG644542 similar to PIR|D84681|D846 hypothetical protein At2g281... 45 7e-05
TC85290 similar to GP|7767653|gb|AAF69150.1| F27F5.2 {Arabidopsi... 39 0.005
TC88253 weakly similar to GP|15810597|gb|AAL07186.1 unknown prot... 39 0.005
AW690594 similar to GP|23498163|emb hypothetical protein {Plasmo... 39 0.005
TC78972 weakly similar to GP|21592657|gb|AAM64606.1 unknown {Ara... 36 0.032
BE204345 similar to GP|15126717|gb| Similar to ectoplacental con... 36 0.032
AJ499220 similar to SP|Q09863|YAF Hypothetical protein C29E6.10c... 35 0.042
TC87847 similar to GP|15929245|gb|AAH15068.1 Similar to adaptor-... 35 0.054
TC77925 similar to GP|18700163|gb|AAL77693.1 AT4g02720/T10P11_1 ... 35 0.054
TC87846 similar to GP|5821151|dbj|BAA83717.1 RNA binding protein... 35 0.054
TC79241 similar to GP|21740501|emb|CAD40825. OSJNBa0006B20.16 {O... 35 0.054
CA920710 similar to PIR|S64314|S643 probable membrane protein YG... 34 0.093
TC77828 similar to GP|16648867|gb|AAL24285.1 HSP associated prot... 34 0.093
CA990541 weakly similar to GP|9758600|dbj gene_id:MDF20.10~pir||... 34 0.12
BG452862 weakly similar to GP|14715258|emb N3 like protein {Medi... 33 0.16
CA990769 weakly similar to GP|21594005|gb| unknown {Arabidopsis ... 33 0.16
AJ499968 weakly similar to GP|23495961|gb| hypothetical protein ... 33 0.16
>BG587694 similar to GP|20260480|gb unknown protein {Arabidopsis thaliana},
partial (36%)
Length = 754
Score = 385 bits (990), Expect = e-107
Identities = 186/187 (99%), Positives = 187/187 (99%)
Frame = +1
Query: 1 MSVTSSSRGRPGTRETSPERTKVWTEPKPKTPRKVSVVYYLSRNGHLEHPHFMEVPLSSP 60
MSVTSSSRGRPGTRETSPERTKVWTEPKPKTPRKVSVVYYLSRNGHLEHPHFMEVPLSSP
Sbjct: 52 MSVTSSSRGRPGTRETSPERTKVWTEPKPKTPRKVSVVYYLSRNGHLEHPHFMEVPLSSP 231
Query: 61 HGLYLKDVIIRLNLLRGKGMPTMYSWSSKRSYKNGFVWHDLTESDFIYPTQGHDYILKGS 120
HGLYLKDVIIRLNLLRGKGMPTMYSWSSKRSYKNGFVWHDLTESDFIYPTQGHDYILKGS
Sbjct: 232 HGLYLKDVIIRLNLLRGKGMPTMYSWSSKRSYKNGFVWHDLTESDFIYPTQGHDYILKGS 411
Query: 121 EILEHETTARPPLEEESDSPVVITRRRNQSWSSIDLNEYRVYKSEPLGENIAADASTQTE 180
EILEHETTARPPLEEESDSPVVITRRRNQSWSSIDLNEYRVYKSEPLGENIAADASTQTE
Sbjct: 412 EILEHETTARPPLEEESDSPVVITRRRNQSWSSIDLNEYRVYKSEPLGENIAADASTQTE 591
Query: 181 EKRRRRR 187
EKRRRR+
Sbjct: 592 EKRRRRK 612
Score = 85.1 bits (209), Expect = 5e-17
Identities = 45/55 (81%), Positives = 46/55 (82%), Gaps = 5/55 (9%)
Frame = +2
Query: 176 STQTEEKRRRR-----RVLREEDEEQEEEEKEKNDEIENEGETENQNQSTELSRE 225
S T +RRRR RVLREEDEEQEEEEKEKNDEIENEGETENQNQSTELSRE
Sbjct: 563 SPPTRRRRRRRNGDGERVLREEDEEQEEEEKEKNDEIENEGETENQNQSTELSRE 727
>BF645395 similar to GP|15808946|gb| auxin-regulated protein {Lycopersicon
esculentum}, partial (19%)
Length = 658
Score = 147 bits (370), Expect = 1e-35
Identities = 85/184 (46%), Positives = 115/184 (62%), Gaps = 14/184 (7%)
Frame = +1
Query: 17 SPERTKVWTEPKPKTPRKVSVVYYLSRNGHLEHPHFMEVPLSSPHG-LYLKDVIIRLNLL 75
S ER + TE + K P V+YYLSRNG LEHPH M V +SS HG L L+DVI RL+ L
Sbjct: 19 SMERKTLETEAEVKVP----VIYYLSRNGQLEHPHLMYVSISSSHGTLCLQDVINRLSFL 186
Query: 76 RGKGMPTMYSWSSKRSYKNGFVWHDLTESDFIYPTQGHDYILKGSEILEHETTARPPLEE 135
RG+G+ MYSWS+KRSY+NGFVW DL+E+D IYP+ H+Y+LKG+ ++E ++ R E
Sbjct: 187 RGQGIANMYSWSTKRSYRNGFVWQDLSENDLIYPSSSHEYVLKGTLLIEEPSSFR-SYET 363
Query: 136 ESDSPVVITRRRNQSWSSIDLN-----------EYRVYKSEPLGE--NIAADASTQTEEK 182
P + + SS+D + +Y++YK+ E A +ASTQTEEK
Sbjct: 364 ILSMPSSKSSNETNNSSSMDADSPSSTAKGSRRDYKLYKATTYREFAEKATNASTQTEEK 543
Query: 183 RRRR 186
R+R
Sbjct: 544 GRQR 555
>CB892180 similar to PIR|D84681|D84 hypothetical protein At2g28150 [imported]
- Arabidopsis thaliana, partial (29%)
Length = 872
Score = 144 bits (364), Expect = 5e-35
Identities = 96/265 (36%), Positives = 129/265 (48%), Gaps = 61/265 (23%)
Frame = +2
Query: 43 RNGHLEHPHFMEVPLSSPHGLYLKDVIIRLNLLRGKGMPTMYSWSSKRSYKNGFVWHDLT 102
R+ LEHPHFMEVP+SSP GL+L+DVI +L+ LRG+GM +YSWS KRSYKNG+VWHDL+
Sbjct: 5 RDRQLEHPHFMEVPISSPDGLFLRDVIHKLDALRGRGMADLYSWSCKRSYKNGYVWHDLS 184
Query: 103 ESDFIYPTQGHDYILKGSEI----------------------LEHETTARPPLEEESDSP 140
E D I P G++Y+LKGSE+ L + R E S S
Sbjct: 185 EDDLILPAHGNEYVLKGSELFCESNSDRFSPISNVKLQSLKRLPEPVSCRSHDEVSSSSS 364
Query: 141 VVITRRRNQSW--------------------------SSIDLNEYRVYKSEPLGENIAAD 174
+ R S S+ L EY++YK++ L AD
Sbjct: 365 SMNEREGRNSQEDEISPRQHTGSSDISPESSDGKSDSQSLPLTEYKIYKTDRL-----AD 529
Query: 175 ASTQTEE------KRRRRRVLREEDEEQEEEEKEKNDEIENEGETENQNQSTELSREEIS 228
ASTQTEE + + R E E E ND E + + +E+ + +S
Sbjct: 530 ASTQTEECAAKHQSQTHKTCTRGVSTEDRSSESECNDICET--QAGHVKDGSEICSDVVS 703
Query: 229 PPPSDSSP-------ETLGTLMKVD 246
PP S+SSP ETL +L++ D
Sbjct: 704 PPFSNSSPSSSGGKTETLESLIRAD 778
>BG644542 similar to PIR|D84681|D846 hypothetical protein At2g28150
[imported] - Arabidopsis thaliana, partial (5%)
Length = 781
Score = 44.7 bits (104), Expect = 7e-05
Identities = 33/101 (32%), Positives = 47/101 (45%), Gaps = 13/101 (12%)
Frame = -3
Query: 329 PDLNRVRLEDKEYFSGSLIETKKVEAP---AFKRSSSYNADSSS----------RLQIME 375
P + +R++DKEYFSGSL+E+K KRSSS+NA+ +S + Q M
Sbjct: 665 PKVTSLRMDDKEYFSGSLVESKMAMEDGHNVLKRSSSFNAERTSAQTYRTSKELKSQNMV 486
Query: 376 HEGDMVRAKCIPRKSKTLLTKKEEGTSSSMDSVSRSQHGSK 416
E +KC R T T K + S +S S+
Sbjct: 485 -ESSSGNSKCNSRSVMTSSTTKSPPSEFSRSPISDGSRSSR 366
>TC85290 similar to GP|7767653|gb|AAF69150.1| F27F5.2 {Arabidopsis
thaliana}, partial (32%)
Length = 1558
Score = 38.5 bits (88), Expect = 0.005
Identities = 22/78 (28%), Positives = 39/78 (49%), Gaps = 6/78 (7%)
Frame = +3
Query: 148 NQSWSSIDLNEYRVY------KSEPLGENIAADASTQTEEKRRRRRVLREEDEEQEEEEK 201
N+S+S EY Y + E E A + EEK +R+ ++E E + E+EK
Sbjct: 672 NESYSKEIFEEYITYLKEKAKEKERKREEEKAKKEKEREEKEKRKEKEKKEKEREREKEK 851
Query: 202 EKNDEIENEGETENQNQS 219
K ++E +++NQ+ +
Sbjct: 852 SKERHKKDESDSDNQDMT 905
Score = 28.9 bits (63), Expect = 3.9
Identities = 16/55 (29%), Positives = 27/55 (49%)
Frame = +3
Query: 180 EEKRRRRRVLREEDEEQEEEEKEKNDEIENEGETENQNQSTELSREEISPPPSDS 234
E K RRR +D + E++EKE++ + G + SR+ + P SD+
Sbjct: 951 ERKHRRRHQSSMDDVDSEKDEKEESRKSRRHGSDRKK------SRKHANSPESDN 1097
>TC88253 weakly similar to GP|15810597|gb|AAL07186.1 unknown protein
{Arabidopsis thaliana}, partial (51%)
Length = 1122
Score = 38.5 bits (88), Expect = 0.005
Identities = 21/50 (42%), Positives = 33/50 (66%)
Frame = +3
Query: 180 EEKRRRRRVLREEDEEQEEEEKEKNDEIENEGETENQNQSTELSREEISP 229
EEK+ + E+ EE +EEEK++++E EN+GE +N+ TE+ R E P
Sbjct: 702 EEKKEEEKKEEEKKEEVKEEEKKESEE-ENKGEDDNK---TEIKRSEYWP 839
>AW690594 similar to GP|23498163|emb hypothetical protein {Plasmodium
falciparum 3D7}, partial (10%)
Length = 633
Score = 38.5 bits (88), Expect = 0.005
Identities = 23/87 (26%), Positives = 40/87 (45%)
Frame = +3
Query: 145 RRRNQSWSSIDLNEYRVYKSEPLGENIAADASTQTEEKRRRRRVLREEDEEQEEEEKEKN 204
+RR + + +E V + E + D + EE+ EEDE +EEE+E+
Sbjct: 30 KRRKVEEHNEEEDEGEVEEEEDEHDEEEEDEHDEEEEEEEEEE---EEDEHDDEEEEEEE 200
Query: 205 DEIENEGETENQNQSTELSREEISPPP 231
+E E E + + + + E+ R P P
Sbjct: 201 EEEEEEEDDDEEGEEDEIDRISTLPDP 281
>TC78972 weakly similar to GP|21592657|gb|AAM64606.1 unknown {Arabidopsis
thaliana}, partial (27%)
Length = 1396
Score = 35.8 bits (81), Expect = 0.032
Identities = 15/38 (39%), Positives = 26/38 (67%)
Frame = -3
Query: 175 ASTQTEEKRRRRRVLREEDEEQEEEEKEKNDEIENEGE 212
+S+ + ++ +++ + ED E+EEEE+EK EIE E E
Sbjct: 413 SSSSYQRRKEKQKTMEVEDREKEEEEEEKEREIERE*E 300
>BE204345 similar to GP|15126717|gb| Similar to ectoplacental cone invasive
trophoblast giant cells extraembryonic ectoderm and,
partial (2%)
Length = 325
Score = 35.8 bits (81), Expect = 0.032
Identities = 16/49 (32%), Positives = 25/49 (50%)
Frame = +1
Query: 190 REEDEEQEEEEKEKNDEIENEGETENQNQSTELSREEISPPPSDSSPET 238
+EED EEEE ++ D +E E E NQ+ + ++ P + P T
Sbjct: 67 QEEDSSSEEEEGDEEDGSSSEDEEEEDNQTQQTPSKDSKPASKNPPPST 213
>AJ499220 similar to SP|Q09863|YAF Hypothetical protein C29E6.10c in
chromosome I. [Fission yeast] {Schizosaccharomyces
pombe}, partial (12%)
Length = 518
Score = 35.4 bits (80), Expect = 0.042
Identities = 18/49 (36%), Positives = 26/49 (52%), Gaps = 7/49 (14%)
Frame = -3
Query: 181 EKRRRRRVLREEDEEQ-------EEEEKEKNDEIENEGETENQNQSTEL 222
E+ RR+ REEDE +E+E E DE E +GE +NQ + +
Sbjct: 513 ERLAERRMQREEDESAMDRGEYYDEDEDEDEDEYEEDGEEDNQTEEQRM 367
>TC87847 similar to GP|15929245|gb|AAH15068.1 Similar to adaptor-related
protein complex AP-3 beta 1 subunit {Mus musculus},
partial (2%)
Length = 931
Score = 35.0 bits (79), Expect = 0.054
Identities = 18/69 (26%), Positives = 34/69 (49%), Gaps = 2/69 (2%)
Frame = +1
Query: 172 AADASTQTEEKRRRRRVLREEDEE--QEEEEKEKNDEIENEGETENQNQSTELSREEISP 229
++D ++ +V+ E+DE+ EEEE ++ D +E E E NQ+ + ++ P
Sbjct: 115 SSDEEQPPSTQKPADKVVNEQDEDSSSEEEEGDEEDGSSSEDEEEEDNQTQQTPSKDSKP 294
Query: 230 PPSDSSPET 238
+ P T
Sbjct: 295 ASKNPPPST 321
>TC77925 similar to GP|18700163|gb|AAL77693.1 AT4g02720/T10P11_1
{Arabidopsis thaliana}, partial (47%)
Length = 1538
Score = 35.0 bits (79), Expect = 0.054
Identities = 30/122 (24%), Positives = 58/122 (46%), Gaps = 19/122 (15%)
Frame = +3
Query: 135 EESDSPVVITRRRN---------QSWSSIDLNEYRVYKSEPLGENIAADASTQTEEKRRR 185
+ESDS + RR++ +S S ++ + + KS E+ + S + + KRRR
Sbjct: 570 KESDSELRKKRRKSYKKSRESDSESESESEVEDRKRRKSRKYSESDSDTNSEEEDRKRRR 749
Query: 186 RR--------VLREEDEEQEEEEKEKNDE--IENEGETENQNQSTELSREEISPPPSDSS 235
RR R++ + +E E +DE ++ G +++ + S+++ S P S+ S
Sbjct: 750 RRKSSRRRRKSSRKKRRCSDSDESETDDESGYDDSGRKRKRSKRSRKSKKKSSEPVSEGS 929
Query: 236 PE 237
E
Sbjct: 930 EE 935
>TC87846 similar to GP|5821151|dbj|BAA83717.1 RNA binding protein {Homo
sapiens}, partial (2%)
Length = 550
Score = 35.0 bits (79), Expect = 0.054
Identities = 18/69 (26%), Positives = 34/69 (49%), Gaps = 2/69 (2%)
Frame = +1
Query: 172 AADASTQTEEKRRRRRVLREEDEE--QEEEEKEKNDEIENEGETENQNQSTELSREEISP 229
++D ++ +V+ E+DE+ EEEE ++ D +E E E NQ+ + ++ P
Sbjct: 97 SSDEEQPPSTQKPADKVVNEQDEDSSSEEEEGDEEDGSSSEDEEEEDNQTQQTPSKDSKP 276
Query: 230 PPSDSSPET 238
+ P T
Sbjct: 277 ASKNPPPST 303
>TC79241 similar to GP|21740501|emb|CAD40825. OSJNBa0006B20.16 {Oryza
sativa}, partial (77%)
Length = 1285
Score = 35.0 bits (79), Expect = 0.054
Identities = 18/35 (51%), Positives = 21/35 (59%)
Frame = +3
Query: 192 EDEEQEEEEKEKNDEIENEGETENQNQSTELSREE 226
EDEE E EE+E D++E E ETE S E EE
Sbjct: 345 EDEEVEVEEEEDVDDVEFEDETEMDEDSVEFYDEE 449
>CA920710 similar to PIR|S64314|S643 probable membrane protein YGR023w -
yeast (Saccharomyces cerevisiae), partial (7%)
Length = 774
Score = 34.3 bits (77), Expect = 0.093
Identities = 14/33 (42%), Positives = 24/33 (72%)
Frame = -2
Query: 191 EEDEEQEEEEKEKNDEIENEGETENQNQSTELS 223
E + E+EEEE++ DE ENE E E++++ E++
Sbjct: 377 EAEAEEEEEEEKDEDEDENEDEDEDEDEDEEVA 279
>TC77828 similar to GP|16648867|gb|AAL24285.1 HSP associated protein like
{Arabidopsis thaliana}, partial (83%)
Length = 1587
Score = 34.3 bits (77), Expect = 0.093
Identities = 31/116 (26%), Positives = 49/116 (41%)
Frame = +3
Query: 125 HETTARPPLEEESDSPVVITRRRNQSWSSIDLNEYRVYKSEPLGENIAADASTQTEEKRR 184
H T P +E + + + + L+ +R Y E LG N+ A +++
Sbjct: 96 HTHTMEPSKLKELKHFIEQVKSNPSTLADPSLSFFRDYL-ESLGANLPESAYSKS----- 257
Query: 185 RRRVLREEDEEQEEEEKEKNDEIENEGETENQNQSTELSREEISPPPSDSSPETLG 240
R V ++D E EEEK K +E+E E E + E E P D P+ +G
Sbjct: 258 -RSVESDDDIEDVEEEKVKVEEVEEEDEIIESDVELEGDTVE----PDDDPPQKMG 410
>CA990541 weakly similar to GP|9758600|dbj
gene_id:MDF20.10~pir||T08929~similar to unknown protein
{Arabidopsis thaliana}, partial (13%)
Length = 712
Score = 33.9 bits (76), Expect = 0.12
Identities = 32/148 (21%), Positives = 58/148 (38%), Gaps = 5/148 (3%)
Frame = +2
Query: 94 NGFVWHDLTESDFIYPTQGHD-----YILKGSEILEHETTARPPLEEESDSPVVITRRRN 148
+GFVWH+ E I + D +L+ S++L+ +E+ + ++
Sbjct: 56 SGFVWHENEEKQMIKVKEKFDKCNKEKLLEFSDLLDVPVGNANTRKEDIIAKLIDFLVAP 235
Query: 149 QSWSSIDLNEYRVYKSEPLGENIAADASTQTEEKRRRRRVLREEDEEQEEEEKEKNDEIE 208
Q S+ L E E + IA ST + RR + E ++K +
Sbjct: 236 QVTKSVLLAEQEKSAKE---KRIAKQGSTGSGTPTSRRSAKSRKKIEDSSIAEKKKTATD 406
Query: 209 NEGETENQNQSTELSREEISPPPSDSSP 236
E E+E ++ E + ++S D P
Sbjct: 407 TESESEKDEENKEENEIDVSDKSEDEKP 490
>BG452862 weakly similar to GP|14715258|emb N3 like protein {Medicago
truncatula}, partial (33%)
Length = 658
Score = 33.5 bits (75), Expect = 0.16
Identities = 15/67 (22%), Positives = 37/67 (54%), Gaps = 9/67 (13%)
Frame = +3
Query: 169 ENIAADASTQTEEKRRRRRVL---------REEDEEQEEEEKEKNDEIENEGETENQNQS 219
EN+ + T +EK+ V+ ++DEE +E+EK+++ +++ + +N+N +
Sbjct: 414 ENVVVPSKTTNDEKKLEVSVVDMVIVEKKEEKQDEEHDEKEKKQDQVTQDKTKVKNENDN 593
Query: 220 TELSREE 226
+++ E
Sbjct: 594 ININKTE 614
>CA990769 weakly similar to GP|21594005|gb| unknown {Arabidopsis thaliana},
partial (20%)
Length = 488
Score = 33.5 bits (75), Expect = 0.16
Identities = 17/65 (26%), Positives = 34/65 (52%)
Frame = +1
Query: 166 PLGENIAADASTQTEEKRRRRRVLREEDEEQEEEEKEKNDEIENEGETENQNQSTELSRE 225
P E + + T+ E+ ++ + EE +E+++EEK + + E + E + + E +E
Sbjct: 136 PKSELLLDTSYTKMGEEAKQEQPKVEEKQEEKKEEKAEGGKEEEKKEESKEEKPEEEKKE 315
Query: 226 EISPP 230
E PP
Sbjct: 316 EPKPP 330
>AJ499968 weakly similar to GP|23495961|gb| hypothetical protein {Plasmodium
falciparum 3D7}, partial (19%)
Length = 342
Score = 33.5 bits (75), Expect = 0.16
Identities = 17/46 (36%), Positives = 25/46 (53%)
Frame = -3
Query: 181 EKRRRRRVLREEDEEQEEEEKEKNDEIENEGETENQNQSTELSREE 226
EK R R REE EEQE KEK ++ E E + + +++E+
Sbjct: 340 EKEERERKEREEREEQERIAKEKEEQERKEREERERLELERIAKEK 203
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.309 0.128 0.359
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,307,366
Number of Sequences: 36976
Number of extensions: 171603
Number of successful extensions: 1328
Number of sequences better than 10.0: 111
Number of HSP's better than 10.0 without gapping: 1202
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1280
length of query: 422
length of database: 9,014,727
effective HSP length: 99
effective length of query: 323
effective length of database: 5,354,103
effective search space: 1729375269
effective search space used: 1729375269
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.7 bits)
S2: 60 (27.7 bits)
Medicago: description of AC139841.8


