

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC139747.4 + phase: 0
(500 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
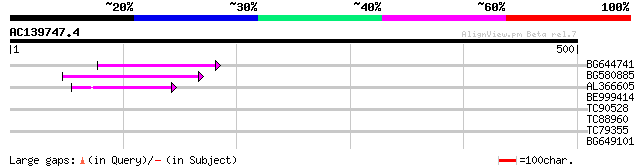
Score E
Sequences producing significant alignments: (bits) Value
BG644741 82 6e-16
BG580885 60 2e-09
AL366605 42 4e-04
BE999414 35 0.087
TC90528 homologue to GP|21592490|gb|AAM64440.1 unknown {Arabidop... 32 0.74
TC88960 similar to GP|21537297|gb|AAM61638.1 DNA-damage inducibl... 30 1.6
TC79355 similar to GP|15081721|gb|AAK82515.1 At1g04990/F13M7_1 {... 29 3.7
BG649101 homologue to GP|6648199|gb|A unknown protein {Arabidops... 28 8.2
>BG644741
Length = 735
Score = 81.6 bits (200), Expect = 6e-16
Identities = 39/109 (35%), Positives = 59/109 (53%)
Frame = -2
Query: 78 LHLFPFSLRDRASAWFHSLQIGSITSWNQMRKAFLSQFFPHSKTAQLRSQITQFTQKDGE 137
L +FP SL A WF L SI +WNQ+R FL++++P SK ++ F GE
Sbjct: 587 LRVFPLSLMGEADIWFTELPYNSIFTWNQLRDVFLARYYPVSKKLNHNDRVNNFVALPGE 408
Query: 138 SLYDAWERFKKMLRLCPHHGFENWLIIHTFYNGLLFSTKMNVDAAAGGA 186
S+ +W+RF LR P+H ++ + FY G + K+ +D AGG+
Sbjct: 407 SVSSSWDRFTSFLRSVPNHRIDDDSLKEYFYRGQDDNNKVVLDTIAGGS 261
>BG580885
Length = 609
Score = 60.1 bits (144), Expect = 2e-09
Identities = 34/127 (26%), Positives = 69/127 (53%), Gaps = 2/127 (1%)
Frame = +3
Query: 47 FFGSPTEDPNLHVSIFLRLSGTLKANQEAVRLHLFPFSLRDRASAWFH-SLQIGSIT-SW 104
F G+P E P H+S F ++ A+ ++ +FP +L + ++ W+ +++ I+ SW
Sbjct: 54 FRGTPNESPITHLSRFNKVCRANNASSVEMQKKIFPVTLEEESALWYDLNIEPYYISLSW 233
Query: 105 NQMRKAFLSQFFPHSKTAQLRSQITQFTQKDGESLYDAWERFKKMLRLCPHHGFENWLII 164
++++ +FL ++ +LRS++ Q + E + + R + +L+ P HG E+ +I
Sbjct: 234 DEIKLSFLQAYYEIEPVEELRSELMGIHQGEKERVRSYFLRLQWILKRWPEHGLEDDVIK 413
Query: 165 HTFYNGL 171
F NGL
Sbjct: 414 GVFVNGL 434
>AL366605
Length = 422
Score = 42.4 bits (98), Expect = 4e-04
Identities = 24/93 (25%), Positives = 49/93 (51%)
Frame = -3
Query: 55 PNLHVSIFLRLSGTLKANQEAVRLHLFPFSLRDRASAWFHSLQIGSITSWNQMRKAFLSQ 114
P H+ ++R G K N +++ +H F SL + A+ W+ SL I +++++ AF S
Sbjct: 291 PQNHIIKYVRKMGNYKDN-DSLMIHCFQDSLMEDAAEWYTSLSKNDIHTFDELAAAFKSH 115
Query: 115 FFPHSKTAQLRSQITQFTQKDGESLYDAWERFK 147
+ +++ R + +QK ES + +R++
Sbjct: 114 YGFNTRLKPNREFLRSLSQKKEESFREYAQRWR 16
>BE999414
Length = 613
Score = 34.7 bits (78), Expect = 0.087
Identities = 21/64 (32%), Positives = 34/64 (52%)
Frame = +3
Query: 52 TEDPNLHVSIFLRLSGTLKANQEAVRLHLFPFSLRDRASAWFHSLQIGSITSWNQMRKAF 111
T DP+ H+ + + T ++ + AV+ LF +LR A WF +L+ SI SW + F
Sbjct: 27 T*DPDEHMEN-IEVVLTYRSVRGAVKCKLFVTTLRRGAMTWFKNLRRNSIGSWGDLCHEF 203
Query: 112 LSQF 115
+ F
Sbjct: 204 TTHF 215
>TC90528 homologue to GP|21592490|gb|AAM64440.1 unknown {Arabidopsis
thaliana}, partial (37%)
Length = 950
Score = 31.6 bits (70), Expect = 0.74
Identities = 18/57 (31%), Positives = 28/57 (48%)
Frame = +1
Query: 396 NAIQLRDGKQLEDTIMKTKTIEGEIESEKQQGEKVIGESDKPIVSPPYKLKIHVPQR 452
NA+ DGK+ + + K + +G++E E G+ S P PP + IHV R
Sbjct: 478 NAVNDSDGKRTKASGNKNEGGDGKVEGEASSGKHAEQSSKPPPSDPPKQDYIHVRAR 648
>TC88960 similar to GP|21537297|gb|AAM61638.1 DNA-damage inducible protein
DDI1-like {Arabidopsis thaliana}, partial (91%)
Length = 1523
Score = 30.4 bits (67), Expect = 1.6
Identities = 22/73 (30%), Positives = 34/73 (46%), Gaps = 9/73 (12%)
Frame = -1
Query: 239 STKVDTLLQKFDKLTVSVVTPALVSPPCEFCGILDHTG-----VECKLGRVAKSPDQNSY 293
ST TLL + L +S P +PPC+ + D TG +E L + S +++
Sbjct: 1268 STSFATLLSNSESLDLSPEAPPEDTPPCDGLLLPDATGSGLPELEASLEYFSSSKNRDGM 1089
Query: 294 TF----GQHTTPP 302
+F G T+PP
Sbjct: 1088 SFSCKNGTETSPP 1050
>TC79355 similar to GP|15081721|gb|AAK82515.1 At1g04990/F13M7_1 {Arabidopsis
thaliana}, partial (42%)
Length = 1627
Score = 29.3 bits (64), Expect = 3.7
Identities = 17/59 (28%), Positives = 26/59 (43%), Gaps = 2/59 (3%)
Frame = +2
Query: 369 VAQKVATSSPTLEVFPGQTEANPK--SLINAIQLRDGKQLEDTIMKTKTIEGEIESEKQ 425
++Q SPTL VF NP+ S + + KQ D + ++ T +S KQ
Sbjct: 1226 ISQNYGLPSPTLSVFDASLLTNPRRLSTVQPAETSPSKQSTDKLQQSDTKAATEDSSKQ 1402
>BG649101 homologue to GP|6648199|gb|A unknown protein {Arabidopsis
thaliana}, partial (3%)
Length = 799
Score = 28.1 bits (61), Expect = 8.2
Identities = 12/35 (34%), Positives = 21/35 (59%)
Frame = +1
Query: 429 KVIGESDKPIVSPPYKLKIHVPQRLAKPNFIVIES 463
K++ +S +++ P +KIH+ Q PNFI+ S
Sbjct: 148 KLVSKSWNTLITSPSFIKIHLNQSSQNPNFILTPS 252
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.316 0.132 0.382
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 14,805,366
Number of Sequences: 36976
Number of extensions: 197361
Number of successful extensions: 1008
Number of sequences better than 10.0: 16
Number of HSP's better than 10.0 without gapping: 1006
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1007
length of query: 500
length of database: 9,014,727
effective HSP length: 100
effective length of query: 400
effective length of database: 5,317,127
effective search space: 2126850800
effective search space used: 2126850800
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 60 (27.7 bits)
Medicago: description of AC139747.4


