

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC139744.6 + phase: 1 /partial
(384 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
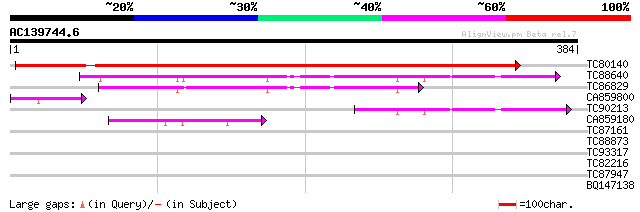
Score E
Sequences producing significant alignments: (bits) Value
TC80140 homologue to GP|21536943|gb|AAM61284.1 putative methioni... 470 e-133
TC88640 similar to GP|9652218|gb|AAF91445.1| putative DNA bindin... 119 2e-27
TC86829 similar to GP|9652218|gb|AAF91445.1| putative DNA bindin... 87 1e-17
CA859800 weakly similar to GP|17473819|gb| putative protein {Ara... 55 3e-08
TC90213 similar to GP|9652218|gb|AAF91445.1| putative DNA bindin... 55 6e-08
CA859180 weakly similar to GP|14532886|gb| putative nuclear DNA-... 50 1e-06
TC87161 similar to GP|15146198|gb|AAK83582.1 F17F16.8/F17F16.8 {... 31 0.70
TC88873 similar to PIR|T02456|T02456 protein kinase homolog F4I1... 30 2.0
TC93317 similar to PIR|T01289|T01289 probable protein kinase At2... 29 3.5
TC82216 similar to GP|16930419|gb|AAL31895.1 AT4g21800/F17L22_26... 28 4.5
TC87947 similar to GP|10178060|dbj|BAB11424. beta-xylosidase {Ar... 28 5.9
BQ147138 similar to PIR|D86390|D86 hypothetical protein AAF98584... 28 5.9
>TC80140 homologue to GP|21536943|gb|AAM61284.1 putative methionine
aminopeptidase {Arabidopsis thaliana}, partial (77%)
Length = 1254
Score = 470 bits (1210), Expect = e-133
Identities = 231/342 (67%), Positives = 273/342 (79%)
Frame = +1
Query: 5 QKKKKNKKKKGKKVQTDPPTIPICELFSDGVYPIGQIMDYPTVNDSRTAKDRFTNEEKRA 64
+KK K+KKKKG QTDPP+IP+ +L+ G +P G+I Y N RT T+EEKR
Sbjct: 244 KKKTKSKKKKGPIEQTDPPSIPVLDLYPSGDFPEGEIQQYKDDNLWRT-----TSEEKRE 408
Query: 65 LDRSESDIYNEVRLAAEAHRQTRKHMQQWIKPGMTMIQICEELENTARKLIKEDGLKAGL 124
L+ + IYN VR AAE HRQ RK+M+ IKPGM M +CE LENT RKLI EDGL+AG+
Sbjct: 409 LELLQKPIYNSVRRAAEVHRQVRKYMKGIIKPGMLMSDLCETLENTVRKLISEDGLQAGI 588
Query: 125 AFPTGCSRNHCAAHYTPNAGDPTVLEYDDVTKIDFGTHINGRIIDCAFTLSFNPKYDKLI 184
AFPTGCS N AAH+TPN+GD TVL+YDDV K+DFGTHI+G I+DCAFT +FNP +D L+
Sbjct: 589 AFPTGCSLNWVAAHWTPNSGDKTVLQYDDVMKLDFGTHIDGYIVDCAFTAAFNPMFDPLL 768
Query: 185 EAVRDATNTGIKAAGIDVPLCEIGAAIQEVMESYEVELDGKTYQVKSIRNLNGHSISPYR 244
EA R+AT TGIK AGIDV LC++GAAIQEVMESYEVE++GK YQVKSIRNLNGHSI Y+
Sbjct: 769 EASREATYTGIKEAGIDVRLCDVGAAIQEVMESYEVEINGKVYQVKSIRNLNGHSIGRYQ 948
Query: 245 IHAGKTVPIVKGGEATVMEENEYYAIETFGSTGRGQVHDDMDCSHYMKNFDAGYMPLRLQ 304
IHAGK+VPIVKGGE T MEE E+YAIETFGSTG+G V +D++CSHYMKNFD G+MPLRL
Sbjct: 949 IHAGKSVPIVKGGEQTKMEEGEFYAIETFGSTGKGYVREDLECSHYMKNFDVGHMPLRLP 1128
Query: 305 SSKSLLSVINKNFSTLAFCKRWLDRAGCTKYQMALKDLCDKG 346
+K LL INKNFSTLAFC+R+LDR G TKY MALK+LCD G
Sbjct: 1129RAKQLLPTINKNFSTLAFCRRYLDRLGETKYLMALKNLCDAG 1254
>TC88640 similar to GP|9652218|gb|AAF91445.1| putative DNA binding protein
{Atriplex hortensis}, partial (91%)
Length = 1514
Score = 119 bits (298), Expect = 2e-27
Identities = 97/348 (27%), Positives = 169/348 (47%), Gaps = 22/348 (6%)
Frame = +1
Query: 48 NDSRTAKDRFTNE--------EKRALDRSESDIYNEVRLAAEAHRQTRKHMQQWIKPGMT 99
N S+ K+ F + +++ LD + D+ + + AAE + + + KP +
Sbjct: 70 NQSKPYKEIFEGQFTMSDDERDEKELDLTSPDVVTKYKSAAEIVNKALQLVISECKPDVK 249
Query: 100 MIQICEELENTAR----KLIK--EDGLKAGLAFPTGCSRNHCAAHYTPNAGDPTVLEYDD 153
++ ICE+ ++ R KL K + ++ G+AFPT S N+ H++P A D TVLE D
Sbjct: 250 IVDICEKGDSFIREQTGKLYKNVKKKIERGVAFPTCISVNNTVCHFSPLASDETVLEVGD 429
Query: 154 VTKIDFGTHINGRIIDCAFT--LSFNPKYDKLIEAVRDATNTGIKAAGIDVPLCEIGAAI 211
+ KID HI+G I A T L P + + + A NT AA + + L + G
Sbjct: 430 ILKIDMACHIDGFIAAVAHTHVLREGPVTGRAADVIA-AANT---AAEVALRLVKPGKKN 597
Query: 212 QEVMESYEVELDGKTYQVKSIRNLNGHSISPYRIHAGKTVPIVKGGEATV----MEENEY 267
Q+V ++ ++ Y + + + H + + I A K V V + V EENE
Sbjct: 598 QDVTDA--IQKVAAAYDCRIVEGVLSHQMKQFVIDANKVVLSVSNPDTRVDDAEFEENEV 771
Query: 268 YAIETFGSTGRG--QVHDDMDCSHYMKNFDAGYMPLRLQSSKSLLSVINKNFSTLAFCKR 325
YAI+ STG G ++ D+ + Y + D Y L++++S+ + S I++NF + F R
Sbjct: 772 YAIDIVASTGEGKPKLLDEKQTTIYKRAVDRSY-NLKMKASRFIYSEISQNFPIMPFSAR 948
Query: 326 WLDRAGCTKYQMALKDLCDKGIVDAYPPLCDVKGCYTAQFEHTIMLRP 373
L+ + ++ L + + ++ YP L + G Y A + T++L P
Sbjct: 949 ALEE---KRARLGLVECMNHELLQPYPVLHEKPGDYVAHIKFTVLLMP 1083
>TC86829 similar to GP|9652218|gb|AAF91445.1| putative DNA binding protein
{Atriplex hortensis}, partial (59%)
Length = 872
Score = 87.0 bits (214), Expect = 1e-17
Identities = 68/233 (29%), Positives = 111/233 (47%), Gaps = 13/233 (5%)
Frame = +3
Query: 61 EKRALDRSESDIYNEVRLAAEAHRQTRKHMQQWIKPGMTMIQICEELENTAR-------K 113
E++ LD + D+ + + AAE + K + KP ++ ICE+ ++ R K
Sbjct: 150 EEKELDLTSPDVVTKYKTAAEIVNKALKLVISECKPKAKVVDICEKGDSFIREQTSNVYK 329
Query: 114 LIKEDGLKAGLAFPTGCSRNHCAAHYTPNAGDPTVLEYDDVTKIDFGTHINGRIIDCAFT 173
+K+ ++ G+AFPT S N+ H++P A D TVL+ D+ KID HI+G I A T
Sbjct: 330 NVKKK-IERGVAFPTCISVNNTICHFSPLASDETVLDEGDIVKIDLACHIDGFIAAVAHT 506
Query: 174 --LSFNPKYDKLIEAVRDATNTGIKAAGIDVPLCEIGAAIQEVMESYEVELDGKTYQVKS 231
L P + + + A NT AA + + L G ++V ++ ++ Y K
Sbjct: 507 HVLQEGPVTGRAADVIA-AANT---AAEVALRLVRPGKKNKDVSDA--IQKVAAAYDCKI 668
Query: 232 IRNLNGHSISPYRIHAGKTVPIVKGGEATV----MEENEYYAIETFGSTGRGQ 280
+ + H + + I K V + + V EENE YAI+ STG G+
Sbjct: 669 VEGVLSHQLKQFVIDGNKVVLSISNPDTRVDDAEFEENEVYAIDIVTSTGEGK 827
>CA859800 weakly similar to GP|17473819|gb| putative protein {Arabidopsis
thaliana}, partial (8%)
Length = 531
Score = 55.5 bits (132), Expect = 3e-08
Identities = 30/68 (44%), Positives = 36/68 (52%), Gaps = 16/68 (23%)
Frame = +2
Query: 1 GGEAQKKKKNKKKKGKKV----------------QTDPPTIPICELFSDGVYPIGQIMDY 44
GGEA KKKK KKK KK Q++PP IP+ + F DG YPIG+I +Y
Sbjct: 323 GGEASKKKKKKKKSKKKTGKVEESGVNDNTSTIQQSEPPRIPVSKFFPDGKYPIGEICEY 502
Query: 45 PTVNDSRT 52
N RT
Sbjct: 503 KDDNLKRT 526
>TC90213 similar to GP|9652218|gb|AAF91445.1| putative DNA binding protein
{Atriplex hortensis}, partial (46%)
Length = 969
Score = 54.7 bits (130), Expect = 6e-08
Identities = 39/153 (25%), Positives = 75/153 (48%), Gaps = 6/153 (3%)
Frame = +3
Query: 234 NLNGHSISPYRIHAGKTVPIVKGGEATV----MEENEYYAIETFGSTGRG--QVHDDMDC 287
+L+ H + + I K V + + V EENE YAI+ STG G ++ D+
Sbjct: 69 SLSLHQLKQFVIDGNKVVLSISNPDTRVDDAEFEENEVYAIDIVTSTGEGKPKLLDEKQT 248
Query: 288 SHYMKNFDAGYMPLRLQSSKSLLSVINKNFSTLAFCKRWLDRAGCTKYQMALKDLCDKGI 347
+ Y + D Y L++++S+ + S I++ F + F R L+ + ++ L + + +
Sbjct: 249 TIYKRAVDKSY-HLKMKASRFIFSEISQKFPIMPFSARALEE---KRARLGLVECMNHEL 416
Query: 348 VDAYPPLCDVKGCYTAQFEHTIMLRPTCKEVVS 380
+ YP L + G + A + T++L P + V+
Sbjct: 417 LQPYPVLHEKPGDFVAHIKFTVLLMPNGSDRVT 515
>CA859180 weakly similar to GP|14532886|gb| putative nuclear DNA-binding
protein G2p {Arabidopsis thaliana}, partial (15%)
Length = 520
Score = 50.1 bits (118), Expect = 1e-06
Identities = 32/115 (27%), Positives = 56/115 (47%), Gaps = 8/115 (6%)
Frame = +1
Query: 68 SESDIYNEVRLAAEAHRQTRKHMQQWIKPGMTMIQIC---EELENTARKLI--KEDGLKA 122
+ SD+ + ++AAE + K + + G ++++C ++L K++ K +
Sbjct: 43 ANSDVVTKYQMAAEVSNRVLKKVIESAVDGAKILELCALGDKLIEDGVKVLYTKNKNMSK 222
Query: 123 GLAFPTGCSRNHCAAHYTPNAGDP---TVLEYDDVTKIDFGTHINGRIIDCAFTL 174
G+AFPT S NH H++P DP L+ D+ KI G I+G A T+
Sbjct: 223 GVAFPTCVSVNHIICHFSPLTSDPEGTEALKNGDLAKIQLGAQIDGYAAIVATTI 387
>TC87161 similar to GP|15146198|gb|AAK83582.1 F17F16.8/F17F16.8 {Arabidopsis
thaliana}, partial (58%)
Length = 607
Score = 31.2 bits (69), Expect = 0.70
Identities = 27/105 (25%), Positives = 50/105 (46%), Gaps = 10/105 (9%)
Frame = +3
Query: 1 GGEAQKKKKNKKKKGKKVQTDPPTIPICELFSDGVYPIGQIMDYPTVND----SRTAK-- 54
GG +KKKKNK+ + + +Q E+ + G + + VND SR K
Sbjct: 285 GGIKKKKKKNKRNQQQFLQATED-----EISAGGSTEQAKDPNDQEVNDESELSREEKPA 449
Query: 55 ---DRFTNEEKRALD-RSESDIYNEVRLAAEAHRQTRKHMQQWIK 95
D T E+R ++ R + D++ ++A ++HR + Q+++
Sbjct: 450 HYDDHLTPAERRYIEQREQLDVHRLAKIANKSHRDRIQDFNQYLR 584
>TC88873 similar to PIR|T02456|T02456 protein kinase homolog F4I18.11 -
Arabidopsis thaliana, partial (19%)
Length = 739
Score = 29.6 bits (65), Expect = 2.0
Identities = 24/71 (33%), Positives = 38/71 (52%), Gaps = 10/71 (14%)
Frame = +2
Query: 182 KLIEAVRDA---TNT-GIKAAGIDVP----LCEIGAAIQEVMESYEVELDGKTYQVKSIR 233
++++A+R + TN+ G+ + G P +C I QEVM V DG TY+ ++IR
Sbjct: 206 RILDAMRASSGGTNSFGLSSEGPHQPPSYFICPI---FQEVMRDPHVAADGFTYEAEAIR 376
Query: 234 NL--NGHSISP 242
+GH SP
Sbjct: 377 GWLDSGHDASP 409
>TC93317 similar to PIR|T01289|T01289 probable protein kinase At2g19410
[imported] - Arabidopsis thaliana, partial (8%)
Length = 608
Score = 28.9 bits (63), Expect = 3.5
Identities = 18/67 (26%), Positives = 34/67 (49%), Gaps = 3/67 (4%)
Frame = +3
Query: 183 LIEAVRDATNTGIKA--AGIDVPLCEIGAAIQEVMESYEVELDGKTYQVKSIRN-LNGHS 239
L++ + D N +K + P +QE+M+ + DG TY+ ++I+ L+ H+
Sbjct: 93 LLKRLSDMANASVKIGRSSAHTPSQYYCPILQEIMDEPYIAGDGFTYEHRAIKAWLSKHN 272
Query: 240 ISPYRIH 246
+SP H
Sbjct: 273 VSPVTKH 293
>TC82216 similar to GP|16930419|gb|AAL31895.1 AT4g21800/F17L22_260
{Arabidopsis thaliana}, partial (31%)
Length = 702
Score = 28.5 bits (62), Expect = 4.5
Identities = 15/51 (29%), Positives = 27/51 (52%)
Frame = +2
Query: 173 TLSFNPKYDKLIEAVRDATNTGIKAAGIDVPLCEIGAAIQEVMESYEVELD 223
T S + D+ +R A + + AGI+ + A+ +E ME+Y+ +LD
Sbjct: 74 TQSLSLALDEFYSNLRSAGVSAVTGAGIEGFFKAVEASAEEYMETYKADLD 226
>TC87947 similar to GP|10178060|dbj|BAB11424. beta-xylosidase {Arabidopsis
thaliana}, partial (55%)
Length = 1662
Score = 28.1 bits (61), Expect = 5.9
Identities = 11/37 (29%), Positives = 20/37 (53%)
Frame = +2
Query: 284 DMDCSHYMKNFDAGYMPLRLQSSKSLLSVINKNFSTL 320
D+DC Y+ + + RL S+ + ++ NF+TL
Sbjct: 158 DLDCGSYLGQYTGRAVKQRLVDEASINNAVSNNFATL 268
>BQ147138 similar to PIR|D86390|D86 hypothetical protein AAF98584.1
[imported] - Arabidopsis thaliana, partial (16%)
Length = 687
Score = 28.1 bits (61), Expect = 5.9
Identities = 23/86 (26%), Positives = 37/86 (42%), Gaps = 2/86 (2%)
Frame = +2
Query: 49 DSRTAKDRFTNEEKRALDRSESDIYNE--VRLAAEAHRQTRKHMQQWIKPGMTMIQICEE 106
+ RT + T EK + + N +R A + HRQ + H+QQ G+ + E+
Sbjct: 119 EKRTIETNKTKNEKDFRKWCKENFINSRSLRHARDIHRQIQGHVQQM---GLKLASCGED 289
Query: 107 LENTARKLIKEDGLKAGLAFPTGCSR 132
+ R L+ L A + P G R
Sbjct: 290 MLQFRRCLVASFFLNAAVKQPDGTYR 367
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.318 0.135 0.405
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,764,768
Number of Sequences: 36976
Number of extensions: 158101
Number of successful extensions: 1351
Number of sequences better than 10.0: 24
Number of HSP's better than 10.0 without gapping: 1056
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1199
length of query: 384
length of database: 9,014,727
effective HSP length: 98
effective length of query: 286
effective length of database: 5,391,079
effective search space: 1541848594
effective search space used: 1541848594
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 59 (27.3 bits)
Medicago: description of AC139744.6


