

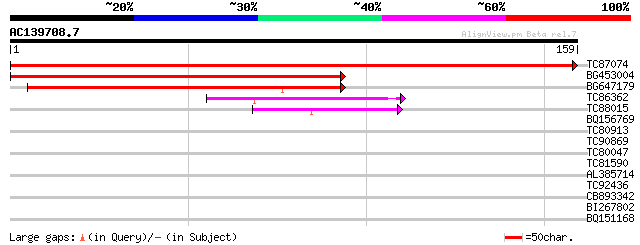
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC139708.7 + phase: 0
(159 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC87074 similar to GP|9758248|dbj|BAB08747.1 gene_id:K3K7.19~unk... 322 5e-89
BG453004 similar to GP|9758248|dbj| gene_id:K3K7.19~unknown prot... 195 6e-51
BG647179 similar to GP|9758248|dbj| gene_id:K3K7.19~unknown prot... 172 5e-44
TC86362 similar to GP|10177062|dbj|BAB10504. gene_id:MKP11.2~unk... 59 6e-10
TC88015 similar to GP|13926206|gb|AAK49581.1 Unknown protein {Ar... 41 2e-04
BQ156769 homologue to GP|13750761|em ORF2 {Rhodococcus erythropo... 35 0.010
TC80913 similar to PIR|T05616|T05616 hypothetical protein F20D10... 30 0.54
TC90869 similar to GP|21536511|gb|AAM60843.1 unknown {Arabidopsi... 28 2.1
TC80047 similar to GP|18087577|gb|AAL58919.1 AT4g00150/F6N15_20 ... 28 2.1
TC81590 similar to GP|6041790|gb|AAF02110.1| putative NADPH-ferr... 26 6.0
AL385714 similar to GP|20259099|gb| putative ubiquitin-conjugati... 26 6.0
TC92436 similar to GP|6862922|gb|AAF30311.1| unknown protein {Ar... 26 7.8
CB893342 weakly similar to PIR|T49117|T49 glucosidase like prote... 26 7.8
BI267802 similar to GP|14517514|gb AT5g52010/MSG15_9 {Arabidopsi... 26 7.8
BQ151168 26 7.8
>TC87074 similar to GP|9758248|dbj|BAB08747.1 gene_id:K3K7.19~unknown
protein {Arabidopsis thaliana}, partial (66%)
Length = 710
Score = 322 bits (824), Expect = 5e-89
Identities = 159/159 (100%), Positives = 159/159 (100%)
Frame = +1
Query: 1 MALQVQASLYTTRLSPTATPTLLRPLSNPSTLKTSFFSRSLNLLLHPNQLQLAYGPPRFT 60
MALQVQASLYTTRLSPTATPTLLRPLSNPSTLKTSFFSRSLNLLLHPNQLQLAYGPPRFT
Sbjct: 88 MALQVQASLYTTRLSPTATPTLLRPLSNPSTLKTSFFSRSLNLLLHPNQLQLAYGPPRFT 267
Query: 61 MRVASKQAYICRDCGYIYNERTAFDKLPDKYFCPVCGAPKRRFKPYATDVNKKANETDVR 120
MRVASKQAYICRDCGYIYNERTAFDKLPDKYFCPVCGAPKRRFKPYATDVNKKANETDVR
Sbjct: 268 MRVASKQAYICRDCGYIYNERTAFDKLPDKYFCPVCGAPKRRFKPYATDVNKKANETDVR 447
Query: 121 KARKAELQRDEAVGKALPIAVAVGVVVLAGLYFYLNSTF 159
KARKAELQRDEAVGKALPIAVAVGVVVLAGLYFYLNSTF
Sbjct: 448 KARKAELQRDEAVGKALPIAVAVGVVVLAGLYFYLNSTF 564
>BG453004 similar to GP|9758248|dbj| gene_id:K3K7.19~unknown protein
{Arabidopsis thaliana}, partial (24%)
Length = 608
Score = 195 bits (496), Expect = 6e-51
Identities = 94/94 (100%), Positives = 94/94 (100%)
Frame = +2
Query: 1 MALQVQASLYTTRLSPTATPTLLRPLSNPSTLKTSFFSRSLNLLLHPNQLQLAYGPPRFT 60
MALQVQASLYTTRLSPTATPTLLRPLSNPSTLKTSFFSRSLNLLLHPNQLQLAYGPPRFT
Sbjct: 80 MALQVQASLYTTRLSPTATPTLLRPLSNPSTLKTSFFSRSLNLLLHPNQLQLAYGPPRFT 259
Query: 61 MRVASKQAYICRDCGYIYNERTAFDKLPDKYFCP 94
MRVASKQAYICRDCGYIYNERTAFDKLPDKYFCP
Sbjct: 260 MRVASKQAYICRDCGYIYNERTAFDKLPDKYFCP 361
>BG647179 similar to GP|9758248|dbj| gene_id:K3K7.19~unknown protein
{Arabidopsis thaliana}, partial (24%)
Length = 620
Score = 172 bits (436), Expect = 5e-44
Identities = 89/116 (76%), Positives = 89/116 (76%), Gaps = 27/116 (23%)
Frame = +3
Query: 6 QASLYTTRLSPTATPTLLRPLSNPSTLKTSFFSRSLNLLLHPNQLQLAYGPPRFTMRVAS 65
QASLYTTRLSPTATPTLLRPLSNPSTLKTSFFSRSLNLLLHPNQLQLAYGPPRFTMRVAS
Sbjct: 3 QASLYTTRLSPTATPTLLRPLSNPSTLKTSFFSRSLNLLLHPNQLQLAYGPPRFTMRVAS 182
Query: 66 KQAYICRDCG---------------------------YIYNERTAFDKLPDKYFCP 94
KQAYICRDCG YIYNERTAFDKLPDKYFCP
Sbjct: 183 KQAYICRDCGLFSTLMLSVSFSFYYLSIYEC***CCRYIYNERTAFDKLPDKYFCP 350
>TC86362 similar to GP|10177062|dbj|BAB10504. gene_id:MKP11.2~unknown
protein {Arabidopsis thaliana}, partial (70%)
Length = 1113
Score = 59.3 bits (142), Expect = 6e-10
Identities = 30/60 (50%), Positives = 35/60 (58%), Gaps = 4/60 (6%)
Frame = +1
Query: 56 PPRFTMRVASKQ----AYICRDCGYIYNERTAFDKLPDKYFCPVCGAPKRRFKPYATDVN 111
PPRF ++ Q +IC DCGYIY +FD PD Y CP C APK+RF Y DVN
Sbjct: 688 PPRFGRKLTETQKARATHICLDCGYIYFLPKSFDDQPDTYSCPQCQAPKKRFAEY--DVN 861
>TC88015 similar to GP|13926206|gb|AAK49581.1 Unknown protein {Arabidopsis
thaliana}, partial (57%)
Length = 741
Score = 41.2 bits (95), Expect = 2e-04
Identities = 19/53 (35%), Positives = 27/53 (50%), Gaps = 11/53 (20%)
Frame = +2
Query: 69 YICRDCGYIYNERTA-----------FDKLPDKYFCPVCGAPKRRFKPYATDV 110
Y CR CGY Y+E F+KLPD + CP CGA + F+ + ++
Sbjct: 341 YECRSCGYKYDEGVGDPSYPIPPGYQFEKLPDDWRCPTCGAAQSFFESKSVEI 499
>BQ156769 homologue to GP|13750761|em ORF2 {Rhodococcus erythropolis},
partial (72%)
Length = 683
Score = 35.4 bits (80), Expect = 0.010
Identities = 17/46 (36%), Positives = 22/46 (46%), Gaps = 11/46 (23%)
Frame = -1
Query: 69 YICRDCGYIYNER-----------TAFDKLPDKYFCPVCGAPKRRF 103
Y C CG+ Y+E T +D +PD + CP CGA K F
Sbjct: 578 YQCAQCGFEYDEAVGWPDDGIEPGTRWDDIPDDWSCPDCGAAKSDF 441
>TC80913 similar to PIR|T05616|T05616 hypothetical protein F20D10.10 -
Arabidopsis thaliana, partial (7%)
Length = 995
Score = 29.6 bits (65), Expect = 0.54
Identities = 29/126 (23%), Positives = 47/126 (37%), Gaps = 16/126 (12%)
Frame = +2
Query: 4 QVQASLYTTRLSPTATPTLLRPLSNPSTLKTSFFSRSLNLLLHPNQLQLAYGPPRFTMRV 63
Q+ S +T +P +P L +S+ S L + + S H + Y + T+R+
Sbjct: 239 QINPSKFTFSTTPPTSPNLHCRISDSSLLDGNKSNNSPKWSSHNSPSSSYYSLLKATLRL 418
Query: 64 ASKQAYICR--------------DCGYIYNER--TAFDKLPDKYFCPVCGAPKRRFKPYA 107
+ IC +C +I++ A K CPVCG +P
Sbjct: 419 SKNLCGICTHSVKTGEGKAIFTAECSHIFHFPCIAAHVKNQQIITCPVCGTNWNDLQPEK 598
Query: 108 TDVNKK 113
T N K
Sbjct: 599 TAENAK 616
>TC90869 similar to GP|21536511|gb|AAM60843.1 unknown {Arabidopsis
thaliana}, partial (78%)
Length = 1104
Score = 27.7 bits (60), Expect = 2.1
Identities = 14/35 (40%), Positives = 19/35 (54%)
Frame = +1
Query: 9 LYTTRLSPTATPTLLRPLSNPSTLKTSFFSRSLNL 43
++TT S T T +PL NP T +T F + NL
Sbjct: 40 VFTTCTSTTEPTTTNQPLHNPKTKRTRFLNPLRNL 144
>TC80047 similar to GP|18087577|gb|AAL58919.1 AT4g00150/F6N15_20
{Arabidopsis thaliana}, partial (30%)
Length = 1357
Score = 27.7 bits (60), Expect = 2.1
Identities = 13/48 (27%), Positives = 25/48 (52%), Gaps = 1/48 (2%)
Frame = +2
Query: 23 LRPLSNPSTLKTSFFSRSLNLLLHPNQLQL-AYGPPRFTMRVASKQAY 69
L P+ NP + + +L L+LH N L A+ P F ++ + +++
Sbjct: 128 LSPIGNPFQRASFYMKEALQLMLHSNGNNLTAFSPISFIFKIGAYKSF 271
>TC81590 similar to GP|6041790|gb|AAF02110.1| putative
NADPH-ferrihemoprotein reductase {Arabidopsis
thaliana}, partial (33%)
Length = 1095
Score = 26.2 bits (56), Expect = 6.0
Identities = 14/40 (35%), Positives = 20/40 (50%)
Frame = +3
Query: 32 LKTSFFSRSLNLLLHPNQLQLAYGPPRFTMRVASKQAYIC 71
LKT FS S + +HPNQ+ L +T K+ +C
Sbjct: 24 LKTRAFSISSSQSVHPNQVHLTVSVVSWTTPYKRKKKGLC 143
>AL385714 similar to GP|20259099|gb| putative ubiquitin-conjugating enzyme E2
{Arabidopsis thaliana}, partial (3%)
Length = 489
Score = 26.2 bits (56), Expect = 6.0
Identities = 13/35 (37%), Positives = 18/35 (51%)
Frame = -1
Query: 11 TTRLSPTATPTLLRPLSNPSTLKTSFFSRSLNLLL 45
+T L + +L P+ PS SFFS + LLL
Sbjct: 270 STSLGKSLESIILNPVEVPSRFSCSFFSPPIGLLL 166
>TC92436 similar to GP|6862922|gb|AAF30311.1| unknown protein {Arabidopsis
thaliana}, partial (56%)
Length = 710
Score = 25.8 bits (55), Expect = 7.8
Identities = 13/27 (48%), Positives = 15/27 (55%)
Frame = +2
Query: 35 SFFSRSLNLLLHPNQLQLAYGPPRFTM 61
S R +LLLHP LQLA P F +
Sbjct: 143 SILRRVPSLLLHPLHLQLALSPSPFLL 223
>CB893342 weakly similar to PIR|T49117|T49 glucosidase like protein -
Arabidopsis thaliana, partial (51%)
Length = 821
Score = 25.8 bits (55), Expect = 7.8
Identities = 13/37 (35%), Positives = 15/37 (40%)
Frame = -3
Query: 46 HPNQLQLAYGPPRFTMRVASKQAYICRDCGYIYNERT 82
H + L Y P F +AYIC C YN T
Sbjct: 432 H*THIPLIYCGPMFNSIPKLSKAYICICCEIFYNSST 322
>BI267802 similar to GP|14517514|gb AT5g52010/MSG15_9 {Arabidopsis thaliana},
partial (35%)
Length = 606
Score = 25.8 bits (55), Expect = 7.8
Identities = 17/62 (27%), Positives = 26/62 (41%), Gaps = 8/62 (12%)
Frame = +1
Query: 80 ERTAFDKLPDKYFCPVCGAP-------KRRFKP-YATDVNKKANETDVRKARKAELQRDE 131
ER P+ Y C VCG K+ FK + + KK N + K +K + ++
Sbjct: 382 ERKGIINPPEPYVCSVCGRKCKTNVDLKKHFKQLHQRERQKKLNRLNSLKGKKRQKYKER 561
Query: 132 AV 133
V
Sbjct: 562 FV 567
>BQ151168
Length = 1011
Score = 25.8 bits (55), Expect = 7.8
Identities = 13/35 (37%), Positives = 18/35 (51%)
Frame = -2
Query: 12 TRLSPTATPTLLRPLSNPSTLKTSFFSRSLNLLLH 46
T +PT + PLS+P +T + RS LL H
Sbjct: 314 TPRAPTPDVSPHAPLSSPQANRTGCYPRSARLLFH 210
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.322 0.136 0.402
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,637,108
Number of Sequences: 36976
Number of extensions: 61789
Number of successful extensions: 421
Number of sequences better than 10.0: 31
Number of HSP's better than 10.0 without gapping: 416
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 419
length of query: 159
length of database: 9,014,727
effective HSP length: 88
effective length of query: 71
effective length of database: 5,760,839
effective search space: 409019569
effective search space used: 409019569
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 54 (25.4 bits)
Medicago: description of AC139708.7


