

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC139706.8 - phase: 0
(324 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
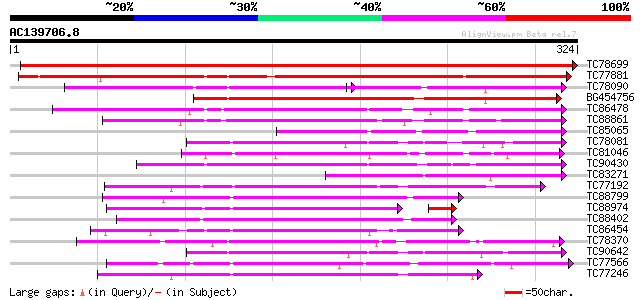
Score E
Sequences producing significant alignments: (bits) Value
TC78699 weakly similar to PIR|G96761|G96761 probable MAP kinase ... 654 0.0
TC77881 homologue to GP|15528439|emb|CAC69137. MEK map kinase ki... 270 4e-73
TC78090 GP|15528441|emb|CAC69138. MAP kinase kinase {Medicago sa... 109 3e-47
BG454756 homologue to GP|12718822|db NQK1 MAPKK {Nicotiana tabac... 166 1e-41
TC86478 similar to GP|3688193|emb|CAA08995.1 MAP3K alpha 1 prote... 137 7e-33
TC88861 weakly similar to GP|17027283|gb|AAL34137.1 putative pro... 127 4e-30
TC85065 similar to GP|1255448|dbj|BAA09057.1 mitogen-activated p... 112 1e-25
TC78081 similar to GP|16904226|gb|AAL30820.1 calcium/calmodulin-... 108 3e-24
TC81046 similar to GP|17529280|gb|AAL38867.1 putative protein ki... 108 4e-24
TC90430 similar to GP|18766642|gb|AAL79042.1 NIMA-related protei... 107 5e-24
TC83271 similar to GP|15528441|emb|CAC69138. MAP kinase kinase {... 105 2e-23
TC77192 similar to SP|P42818|KPK1_ARATH Serine/threonine-protein... 105 2e-23
TC88799 similar to PIR|C71408|C71408 probable protein kinase - A... 105 2e-23
TC88974 similar to GP|3819699|emb|CAA08758.1 BnMAP4K alpha2 {Bra... 100 3e-23
TC88402 similar to GP|20804521|dbj|BAB92215. putative CRK1 prote... 103 9e-23
TC86454 homologue to GP|4567091|gb|AAD23582.1| SNF-1-like serine... 100 6e-22
TC78370 similar to GP|5001828|gb|AAD37165.1| 3-phosphoinositide-... 99 2e-21
TC90642 similar to GP|16904222|gb|AAL30818.1 calcium/calmodulin-... 99 2e-21
TC77566 similar to GP|310580|gb|AAA34017.1|| protein kinase 2 {G... 96 1e-20
TC77246 similar to PIR|C71408|C71408 probable protein kinase - A... 96 1e-20
>TC78699 weakly similar to PIR|G96761|G96761 probable MAP kinase T9L24.32
[imported] - Arabidopsis thaliana, partial (71%)
Length = 1119
Score = 654 bits (1686), Expect = 0.0
Identities = 318/318 (100%), Positives = 318/318 (100%)
Frame = +3
Query: 7 RNSPKLRLPEISDHRPRFPVPLPANIYKQPSTSATTASVAGGDNISAGDFEKLSVLGHGN 66
RNSPKLRLPEISDHRPRFPVPLPANIYKQPSTSATTASVAGGDNISAGDFEKLSVLGHGN
Sbjct: 54 RNSPKLRLPEISDHRPRFPVPLPANIYKQPSTSATTASVAGGDNISAGDFEKLSVLGHGN 233
Query: 67 GGTVYKVRHKLTSIIYALKINHYDSDPTTRRRALTEVNILRRATDCTNVVKYHGSFEKPT 126
GGTVYKVRHKLTSIIYALKINHYDSDPTTRRRALTEVNILRRATDCTNVVKYHGSFEKPT
Sbjct: 234 GGTVYKVRHKLTSIIYALKINHYDSDPTTRRRALTEVNILRRATDCTNVVKYHGSFEKPT 413
Query: 127 GDVCILMEYMDSGSLETALKTTGTFSESKLSTVARDILNGLTYLHARNIAHRDIKPSNIL 186
GDVCILMEYMDSGSLETALKTTGTFSESKLSTVARDILNGLTYLHARNIAHRDIKPSNIL
Sbjct: 414 GDVCILMEYMDSGSLETALKTTGTFSESKLSTVARDILNGLTYLHARNIAHRDIKPSNIL 593
Query: 187 VNIKNEVKIADFGVSKFMGRTLEACNSYVGTCAYMSPERFDPEVYGGNYNGFSADIWSLG 246
VNIKNEVKIADFGVSKFMGRTLEACNSYVGTCAYMSPERFDPEVYGGNYNGFSADIWSLG
Sbjct: 594 VNIKNEVKIADFGVSKFMGRTLEACNSYVGTCAYMSPERFDPEVYGGNYNGFSADIWSLG 773
Query: 247 LTLFELYVGYFPFLQSGQRPDWASLMCAICFSDPPSLPETASSEFRNFVECCLKKESGER 306
LTLFELYVGYFPFLQSGQRPDWASLMCAICFSDPPSLPETASSEFRNFVECCLKKESGER
Sbjct: 774 LTLFELYVGYFPFLQSGQRPDWASLMCAICFSDPPSLPETASSEFRNFVECCLKKESGER 953
Query: 307 WSAAQLLTHPFLCKDMES 324
WSAAQLLTHPFLCKDMES
Sbjct: 954 WSAAQLLTHPFLCKDMES 1007
>TC77881 homologue to GP|15528439|emb|CAC69137. MEK map kinase kinsae
{Medicago sativa subsp. x varia}, complete
Length = 1546
Score = 270 bits (691), Expect = 4e-73
Identities = 143/319 (44%), Positives = 204/319 (63%), Gaps = 3/319 (0%)
Frame = +2
Query: 6 RRNSPKLRLPEISDHRPRFPVPLPANIYKQPSTSATTASVAGGDN---ISAGDFEKLSVL 62
RR+ L LP+ D P+PLP + + + + G I + E+L+ +
Sbjct: 311 RRDHLTLPLPQ-RDTNLAVPLPLPPSGGSGGGGNGSGSGSGGASQQLVIPFSELERLNRI 487
Query: 63 GHGNGGTVYKVRHKLTSIIYALKINHYDSDPTTRRRALTEVNILRRATDCTNVVKYHGSF 122
G G+GGTVYKV H++ YALK+ + + + RR+ E+ ILR D NVVK H +
Sbjct: 488 GSGSGGTVYKVVHRINGRAYALKVIYGHHEESVRRQIHREIQILRDVDD-VNVVKCHEMY 664
Query: 123 EKPTGDVCILMEYMDSGSLETALKTTGTFSESKLSTVARDILNGLTYLHARNIAHRDIKP 182
+ ++ +L+EYMD GSLE E++L+ VAR IL GL YLH R+I HRDIKP
Sbjct: 665 DH-NAEIQVLLEYMDGGSLEGKHIP----QENQLADVARQILRGLAYLHRRHIVHRDIKP 829
Query: 183 SNILVNIKNEVKIADFGVSKFMGRTLEACNSYVGTCAYMSPERFDPEVYGGNYNGFSADI 242
SN+L+N + +VKIADFGV + + +T++ CNS VGT AYMSPER + ++ G Y+ ++ DI
Sbjct: 830 SNLLINSRKQVKIADFGVGRILNQTMDPCNSSVGTIAYMSPERINTDINDGQYDAYAGDI 1009
Query: 243 WSLGLTLFELYVGYFPFLQSGQRPDWASLMCAICFSDPPSLPETASSEFRNFVECCLKKE 302
WSLG+++ E Y+G FPF G++ DWASLMCAIC S PP P TAS EFR+FV CL+++
Sbjct: 1010WSLGVSILEFYMGRFPF-AVGRQGDWASLMCAICMSQPPEAPTTASPEFRDFVSRCLQRD 1186
Query: 303 SGERWSAAQLLTHPFLCKD 321
RW+A++LL+HPFL ++
Sbjct: 1187PSRRWTASRLLSHPFLVRN 1243
>TC78090 GP|15528441|emb|CAC69138. MAP kinase kinase {Medicago sativa subsp.
x varia}, complete
Length = 1677
Score = 109 bits (273), Expect(2) = 3e-47
Identities = 65/168 (38%), Positives = 93/168 (54%), Gaps = 1/168 (0%)
Frame = +2
Query: 32 IYKQPSTSATTASVAGGDNISAGDFEKLSVLGHGNGGTVYKVRHKLTSIIYALKINHYDS 91
I + A A + +S D + + V+G GNGG V V+HK T+ +ALKI +
Sbjct: 356 IVSETEVEAPPPIKATDNQLSLADIDIVKVVGKGNGGVVQLVQHKWTNQFFALKIIQMNI 535
Query: 92 DPTTRRRALTEVNILRRATDCTNVVKYHGSFEKPTGDVCILMEYMDSGSLETALKTTGTF 151
+ + R++ E+ I +A C VV + SF G + I++EYMD GS+ LK T
Sbjct: 536 EESVRKQIAKELKI-NQAAQCPYVVVCYQSFYD-NGVISIILEYMDGGSMADLLKKVKTI 709
Query: 152 SESKLSTVARDILNGLTYL-HARNIAHRDIKPSNILVNIKNEVKIADF 198
E LS + + +L GL YL H R+I HRD+KPSN+L+N EVKI F
Sbjct: 710 PEPYLSAICKQVLKGLIYLHHERHIIHRDLKPSNLLINHTGEVKIT*F 853
Score = 96.7 bits (239), Expect(2) = 3e-47
Identities = 56/130 (43%), Positives = 74/130 (56%), Gaps = 4/130 (3%)
Frame = +3
Query: 193 VKIADFGVSKFMGRTLEACNSYVGTCAYMSPERFDPEVYGGNYNGFSADIWSLGLTLFEL 252
+++ DFGVS M T N+++GT YMSPER + G NY +DIWSLGL L E
Sbjct: 837 LRLPDFGVSAIMESTSGQANTFIGTYNYMSPERINGSQRGYNY---KSDIWSLGLILLEC 1007
Query: 253 YVGYFPFLQSGQRPDWAS---LMCAICFSDPPSLP-ETASSEFRNFVECCLKKESGERWS 308
+G FP+ Q W S L+ I PPS P E SSEF +F+ CL+K+ G R S
Sbjct: 1008AMGRFPYTPPDQSERWESIFELIETIVDKPPPSAPSEQFSSEFCSFISACLQKDPGSRLS 1187
Query: 309 AAQLLTHPFL 318
A +L+ PF+
Sbjct: 1188AQELMELPFI 1217
>BG454756 homologue to GP|12718822|db NQK1 MAPKK {Nicotiana tabacum}, partial
(60%)
Length = 648
Score = 166 bits (419), Expect = 1e-41
Identities = 93/215 (43%), Positives = 130/215 (60%), Gaps = 5/215 (2%)
Frame = +2
Query: 106 LRRATDCTNVVKYHGSFEKPTGDVCILMEYMDSGSLETALKTTGTFSESKLSTVARDILN 165
+ +A+ C +VV + SF G + +++EYMD GSL ++ T E L+ V + +L
Sbjct: 20 INQASQCPHVVVCYHSFYN-NGVISLVLEYMDRGSLVDVIRQVNTILEPYLAVVCKQVLQ 196
Query: 166 GLTYLH-ARNIAHRDIKPSNILVNIKNEVKIADFGVSKFMGRTLEACNSYVGTCAYMSPE 224
GL YLH R++ HRDIKPSN+LVN K EVKI DFGVS + T+ +++VGT YMSPE
Sbjct: 197 GLVYLHNERHVIHRDIKPSNLLVNHKGEVKITDFGVSAMLASTMGQRDTFVGTYNYMSPE 376
Query: 225 RFDPEVYGGNYNGFSADIWSLGLTLFELYVGYFPFLQSGQRPDWAS---LMCAICFSDPP 281
R Y +S DIWSLG+ + E +G FP++QS + W S L+ AI S PP
Sbjct: 377 RISGSTY-----DYSCDIWSLGMVVLECAIGRFPYIQSEDQQAWPSFYELLQAIVESPPP 541
Query: 282 SL-PETASSEFRNFVECCLKKESGERWSAAQLLTH 315
S P+ S EF +FV C+KK+ ER ++ +LL H
Sbjct: 542 SAPPDQFSPEFCSFVSSCIKKDPRERSTSLELLDH 646
>TC86478 similar to GP|3688193|emb|CAA08995.1 MAP3K alpha 1 protein kinase
{Brassica napus}, partial (52%)
Length = 2266
Score = 137 bits (344), Expect = 7e-33
Identities = 97/301 (32%), Positives = 155/301 (51%), Gaps = 7/301 (2%)
Frame = +2
Query: 25 PVPLPANIYKQPSTS-ATTASVAGGDNISAGDFEKLSVLGHGNGGTVYKVRHKLTSIIYA 83
P+PLP PS++ + T S + + ++K +LG G G VY + + A
Sbjct: 875 PLPLPPGSPTSPSSALSNTRSPFENSSPNLSKWKKGKLLGRGTFGHVYLGFNSENGQMCA 1054
Query: 84 LKINHYDSDPTTRRRALT----EVNILRRATDCTNVVKYHGSFEKPTGDVCILMEYMDSG 139
+K D + L E+++L + + N+V+Y GS E + + +EY+ G
Sbjct: 1055 IKEVRVGCDDQNSKECLKQLHQEIDLLNQLSH-PNIVQYLGS-ELGEESLSVYLEYVSGG 1228
Query: 140 SLETALKTTGTFSESKLSTVARDILNGLTYLHARNIAHRDIKPSNILVNIKNEVKIADFG 199
S+ L+ G F E + R I++GL YLH RN HRDIK +NILV+ E+K+ADFG
Sbjct: 1229 SIHKLLQEYGPFKEPVIQNYTRQIVSGLAYLHGRNTVHRDIKGANILVDPNGEIKLADFG 1408
Query: 200 VSKFMGRTLEACNSYVGTCAYMSPERFDPEVYGGNYNGFS--ADIWSLGLTLFELYVGYF 257
++K + + + S+ G+ +M+PE N NG+S DIWSLG TL E+
Sbjct: 1409 MAKHI-TSAASMLSFKGSPYWMAPE------VVMNTNGYSLPVDIWSLGCTLIEMAASKP 1567
Query: 258 PFLQSGQRPDWASLMCAICFSDPPSLPETASSEFRNFVECCLKKESGERWSAAQLLTHPF 317
P+ Q A++ D P +PE S++ +NF+ CL+++ R +A +LL HPF
Sbjct: 1568 PW---SQYEGVAAIFKIGNSKDMPIIPEHLSNDAKNFIMLCLQRDPSARPTAQKLLEHPF 1738
Query: 318 L 318
+
Sbjct: 1739 I 1741
>TC88861 weakly similar to GP|17027283|gb|AAL34137.1 putative protein kinase
{Oryza sativa (japonica cultivar-group)}, partial (34%)
Length = 1847
Score = 127 bits (320), Expect = 4e-30
Identities = 97/280 (34%), Positives = 152/280 (53%), Gaps = 15/280 (5%)
Frame = +1
Query: 54 GDFEKLSVLGHGNGGTVYKVRHKLTSIIYALK-INHYDSDPTTR---RRALTEVNILRRA 109
G ++K ++G G+ G+VY + T ALK ++ DP + ++ E+ IL +
Sbjct: 1018 GHWQKGKLIGRGSFGSVYHATNLETGASCALKEVDLVPDDPKSTDCIKQLDQEIRILGQL 1197
Query: 110 TDCTNVVKYHGSFEKPTGD-VCILMEYMDSGSLETALKT-TGTFSESKLSTVARDILNGL 167
N+V+Y+GS + GD +CI MEY+ GSL+ ++ G +ES + R IL+GL
Sbjct: 1198 HH-PNIVEYYGS--EVVGDRLCIYMEYVHPGSLQKFMQDHCGVMTESVVRNFTRHILSGL 1368
Query: 168 TYLHARNIAHRDIKPSNILVNIKNEVKIADFGVSKFM-GRTLEACNSYVGTCAYMSPE-- 224
YLH+ HRDIK +N+LV+ VK+ADFGVSK + ++ E S G+ +M+PE
Sbjct: 1369 AYLHSTKTIHRDIKGANLLVDASGIVKLADFGVSKILTEKSYEL--SLKGSPYWMAPELM 1542
Query: 225 ------RFDPEVYGGNYNGFSADIWSLGLTLFELYVGYFPFLQSGQRPDWASLMCAICFS 278
+P V + DIWSLG T+ E+ G P+ + P ++ + S
Sbjct: 1543 MAAMKNETNPTV------AMAVDIWSLGCTIIEMLTGKPPW---SEFPGHQAMFKVLHRS 1695
Query: 279 DPPSLPETASSEFRNFVECCLKKESGERWSAAQLLTHPFL 318
P +P+T S E ++F+E C ++ +R SAA LLTHPF+
Sbjct: 1696 --PDIPKTLSPEGQDFLEQCFQRNPADRPSAAVLLTHPFV 1809
>TC85065 similar to GP|1255448|dbj|BAA09057.1 mitogen-activated protein
kinase {Arabidopsis thaliana}, partial (26%)
Length = 789
Score = 112 bits (281), Expect = 1e-25
Identities = 70/166 (42%), Positives = 95/166 (57%)
Frame = +2
Query: 153 ESKLSTVARDILNGLTYLHARNIAHRDIKPSNILVNIKNEVKIADFGVSKFMGRTLEACN 212
+S++S R IL+GL YLH RNI HRDIK +NILV+ VK+ADFG+S + L
Sbjct: 17 DSQVSAYTRQILHGLKYLHDRNIVHRDIKCANILVDANGSVKVADFGLS*AI--KLNDVK 190
Query: 213 SYVGTCAYMSPERFDPEVYGGNYNGFSADIWSLGLTLFELYVGYFPFLQSGQRPDWASLM 272
S GT +M+PE +V G G ADIWSLG T+ E+ G P+ + S M
Sbjct: 191 SCQGTAFWMAPEVVRGKVKG---YGLPADIWSLGCTVLEMLTGQVPYAPM----ECISAM 349
Query: 273 CAICFSDPPSLPETASSEFRNFVECCLKKESGERWSAAQLLTHPFL 318
I + P +P+T S + R+F+ CLK +R +AAQLL H F+
Sbjct: 350 FRIGKGELPPVPDTLSRDARDFILQCLKVNPDDRPTAAQLLDHKFV 487
>TC78081 similar to GP|16904226|gb|AAL30820.1 calcium/calmodulin-dependent
protein kinase CaMK3 {Nicotiana tabacum}, partial (58%)
Length = 1066
Score = 108 bits (270), Expect = 3e-24
Identities = 82/232 (35%), Positives = 121/232 (51%), Gaps = 15/232 (6%)
Frame = +2
Query: 102 EVNILRRATDCTNVVKYHGSFEKPTGDVCILMEYMDSGSL-ETALKTTGTFSESKLSTVA 160
EV ILR +N+VK++ +FE +V I+ME + G L + L G +SE V
Sbjct: 83 EVKILRALNGHSNLVKFYDAFEDQE-NVYIVMELCEGGELLDMILSRGGKYSEDDAKAVM 259
Query: 161 RDILNGLTYLHARNIAHRDIKPSNILVNIK---NEVKIADFGVSKFMGRTLEACNSYVGT 217
ILN + + H + + HRD+KP N L K +E+K DFG+S F+ R E N VG+
Sbjct: 260 VQILNVVAFCHLQGVVHRDLKPENFLYTTKDESSELKAIDFGLSDFV-RPDERLNDIVGS 436
Query: 218 CAYMSPERFDPEVYGGNYNGFSADIWSLGLTLFELYVGYFPFLQSGQRPDWA----SLMC 273
Y++ PEV +Y+ AD+WS+G+ + L G RP WA +
Sbjct: 437 AYYVA-----PEVLHRSYS-TEADVWSIGVIAY--------ILLCGSRPFWARTESGIFR 574
Query: 274 AICFSDP-------PSLPETASSEFRNFVECCLKKESGERWSAAQLLTHPFL 318
A+ +DP PSL SSE ++FV+ L K+ +R SAAQ L+HP++
Sbjct: 575 AVLKADPGFDEGPWPSL----SSEAKDFVKRLLNKDPRKRISAAQALSHPWI 718
>TC81046 similar to GP|17529280|gb|AAL38867.1 putative protein kinase
{Arabidopsis thaliana}, partial (44%)
Length = 921
Score = 108 bits (269), Expect = 4e-24
Identities = 80/234 (34%), Positives = 123/234 (52%), Gaps = 15/234 (6%)
Frame = +2
Query: 99 ALTEVNILRRAT------DCTNVVKYHGSFEKPTGDVCILMEYMDSGSLETALKTTGT-- 150
A+T ++ +RR D N+++ H SF + ++M +M GS +K++
Sbjct: 2 AITNLDGIRREVQTMSLIDHPNLLRAHCSFTAGHS-LWVVMPFMSGGSCLHIMKSSFPEG 178
Query: 151 FSESKLSTVARDILNGLTYLHARNIAHRDIKPSNILVNIKNEVKIADFGVSKFM---GRT 207
F E ++TV R++L L YLHA HRD+K NIL++ VK+ADFGVS M G
Sbjct: 179 FDEPVIATVLREVLKALVYLHAHGHIHRDVKAGNILLDANGSVKMADFGVSACMFDTGDR 358
Query: 208 LEACNSYVGTCAYMSPERFDPEVYGGNYNGFSADIWSLGLTLFELYVGYFPFLQSGQRPD 267
+ N++VGT +M+PE +++G + F ADIWS G+T EL G+ PF + P
Sbjct: 359 QRSRNTFVGTPCWMAPEVMQ-QLHGYD---FKADIWSFGITALELAHGHAPF---SKYPP 517
Query: 268 WASLMCAICFSDPPSL----PETASSEFRNFVECCLKKESGERWSAAQLLTHPF 317
L+ + + PP L + S F+ V CL K+ +R S+ +LL H F
Sbjct: 518 MKVLLMTL-QNAPPGLDYERDKRFSKSFKELVATCLVKDPKKRPSSEKLLKHHF 676
>TC90430 similar to GP|18766642|gb|AAL79042.1 NIMA-related protein kinase
{Populus x canescens}, partial (45%)
Length = 976
Score = 107 bits (268), Expect = 5e-24
Identities = 84/249 (33%), Positives = 125/249 (49%), Gaps = 3/249 (1%)
Frame = +1
Query: 73 VRHKLTSIIYALK-INHYDSDPTTRRRALTEVNILRRATDCTNVVKYHGSFEKPTGDVCI 131
VRHK Y LK I TRR A E+ ++ + + +V+Y S+ + VCI
Sbjct: 136 VRHKHEKKKYVLKKIRLARQTDRTRRSAHQEMELISKVRN-PFIVEYKDSWVEKGCFVCI 312
Query: 132 LMEYMDSGSL-ETALKTTGT-FSESKLSTVARDILNGLTYLHARNIAHRDIKPSNILVNI 189
++ Y + G + ET K G F E KL +L L YLH +I HRD+K SNI +
Sbjct: 313 IIGYCEGGDMAETVKKANGVNFPEEKLCKWLVQLLMALDYLHVNHILHRDVKCSNIFLTK 492
Query: 190 KNEVKIADFGVSKFMGRTLEACNSYVGTCAYMSPERFDPEVYGGNYNGFSADIWSLGLTL 249
++++ DFG++K + + + +S VGT +YM PE YG +DIWSLG +
Sbjct: 493 NQDIRLGDFGLAKLL-TSDDLASSIVGTPSYMCPELLADIPYGS-----KSDIWSLGCCV 654
Query: 250 FELYVGYFPFLQSGQRPDWASLMCAICFSDPPSLPETASSEFRNFVECCLKKESGERWSA 309
+E+ + P ++ D +L+ I S LP SS FR V+ L+K R +A
Sbjct: 655 YEM-AAHRPAFKAF---DIQALIHKINKSIVSPLPTMYSSAFRGLVKSMLRKNPELRPTA 822
Query: 310 AQLLTHPFL 318
+LL HP L
Sbjct: 823 GELLNHPHL 849
>TC83271 similar to GP|15528441|emb|CAC69138. MAP kinase kinase {Medicago
sativa subsp. x varia}, partial (42%)
Length = 683
Score = 105 bits (263), Expect = 2e-23
Identities = 61/142 (42%), Positives = 77/142 (53%), Gaps = 4/142 (2%)
Frame = +1
Query: 181 KPSNILVNIKNEVKIADFGVSKFMGRTLEACNSYVGTCAYMSPERFDPEVYGGNYNGFSA 240
KPSNIL+N + EVKI DFGVS M T N+++GT YMSPER YN + +
Sbjct: 10 KPSNILINHRGEVKITDFGVSAIMESTSGQANTFIGTYNYMSPERIKASDSEQGYN-YKS 186
Query: 241 DIWSLGLTLFELYVGYFPFLQSGQRPDWASLMC---AICFSDPPSL-PETASSEFRNFVE 296
DIWSLGL L + G FP+ W +L AI PS P+ S EF +F+
Sbjct: 187 DIWSLGLMLLKCATGKFPYTPPDNSEGWENLFLLIEAIVEKPSPSAPPDECSPEFCSFIS 366
Query: 297 CCLKKESGERWSAAQLLTHPFL 318
CL+K +R S LL HPF+
Sbjct: 367 ACLQKNPRDRPSTRNLLRHPFV 432
>TC77192 similar to SP|P42818|KPK1_ARATH Serine/threonine-protein kinase
AtPK1/AtPK6 (EC 2.7.1.-). [Mouse-ear cress] {Arabidopsis
thaliana}, partial (76%)
Length = 1992
Score = 105 bits (263), Expect = 2e-23
Identities = 79/254 (31%), Positives = 127/254 (49%), Gaps = 2/254 (0%)
Frame = +3
Query: 55 DFEKLSVLGHGNGGTVYKVRHKLTSIIYALKINHYDS--DPTTRRRALTEVNILRRATDC 112
DFE L V+G G VY+VR K TS IYA+K+ D + E IL + +
Sbjct: 765 DFEVLKVVGQGAFAKVYQVRKKGTSEIYAMKVMRKDKIMEKNHAEYMKAEREILTKI-EH 941
Query: 113 TNVVKYHGSFEKPTGDVCILMEYMDSGSLETALKTTGTFSESKLSTVARDILNGLTYLHA 172
VV+ SF+ + +++++++ G L L G F E A +I++ +++LH+
Sbjct: 942 PFVVQLRYSFQTKYR-LYLVLDFVNGGHLFFQLYHQGLFREDLARIYAAEIVSAVSHLHS 1118
Query: 173 RNIAHRDIKPSNILVNIKNEVKIADFGVSKFMGRTLEACNSYVGTCAYMSPERFDPEVYG 232
+ I HRD+KP NIL++ V + DFG++K + + NS GT YM+ PE+
Sbjct: 1119 KGIMHRDLKPENILMDADGHVMLTDFGLAKKFEESTRS-NSLCGTAEYMA-----PEIIL 1280
Query: 233 GNYNGFSADIWSLGLTLFELYVGYFPFLQSGQRPDWASLMCAICFSDPPSLPETASSEFR 292
G + +AD WS+G+ LFE+ G PF + ++ D LP SS+
Sbjct: 1281 GKGHDKAADWWSVGILLFEMLTGKPPFCGGNCQKIQQKIV-----KDKIKLPGFLSSDAH 1445
Query: 293 NFVECCLKKESGER 306
++ L KE+ +R
Sbjct: 1446 ALLKGLLNKEAPKR 1487
>TC88799 similar to PIR|C71408|C71408 probable protein kinase - Arabidopsis
thaliana, partial (57%)
Length = 1648
Score = 105 bits (262), Expect = 2e-23
Identities = 66/210 (31%), Positives = 105/210 (49%), Gaps = 4/210 (1%)
Frame = +1
Query: 54 GDFEKLSVLGHGNGGTVYKVRHKLTSI-IYALKI--NHYDSDPTTRRRALTEVNILRRAT 110
G ++ +LG G+ VYK L + A+KI + D R + E++ +RR
Sbjct: 121 GKYQLTRLLGRGSFAKVYKAHSLLDNANTVAVKIIDKSKNVDAAMEPRIIREIDAMRRLQ 300
Query: 111 DCTNVVKYHGSFEKPTGDVCILMEYMDSGSLETALKTTGTFSESKLSTVARDILNGLTYL 170
N++K H T + +++E+ G L +AL G FSES + +++ L +
Sbjct: 301 HHPNILKIHEVMATKT-KIHLIVEFAAGGELFSALSRRGRFSESTARFYFQQLVSALRFC 477
Query: 171 HARNIAHRDIKPSNILVNIKNEVKIADFGVSKFMGRTLEA-CNSYVGTCAYMSPERFDPE 229
H +AHRD+KP N+L++ +K++DFG+S + ++ GT AY +PE
Sbjct: 478 HRNGVAHRDLKPQNLLLDADGNLKVSDFGLSALPEHLKDGLLHTACGTPAYTAPEILR-- 651
Query: 230 VYGGNYNGFSADIWSLGLTLFELYVGYFPF 259
G Y+G AD WS GL L+ L GY PF
Sbjct: 652 -RSGGYDGSKADAWSCGLILYVLLAGYLPF 738
>TC88974 similar to GP|3819699|emb|CAA08758.1 BnMAP4K alpha2 {Brassica
napus}, partial (34%)
Length = 851
Score = 100 bits (249), Expect(2) = 3e-23
Identities = 58/169 (34%), Positives = 94/169 (55%)
Frame = +3
Query: 56 FEKLSVLGHGNGGTVYKVRHKLTSIIYALKINHYDSDPTTRRRALTEVNILRRATDCTNV 115
F L ++G G+ G VYK K + A+K+ + E+++L + C +
Sbjct: 189 FTSLELIGQGSFGDVYKGFDKELNKEVAIKVIDLEESEDDIDDIQKEISVLSQCR-CPYI 365
Query: 116 VKYHGSFEKPTGDVCILMEYMDSGSLETALKTTGTFSESKLSTVARDILNGLTYLHARNI 175
+Y+GSF T + I+MEYM GS+ L++ E ++ + RD+L+ + YLH
Sbjct: 366 TEYYGSFLNQT-KLWIIMEYMAGGSVADLLQSGPPLDEMSIAYILRDLLHAVDYLHNEGK 542
Query: 176 AHRDIKPSNILVNIKNEVKIADFGVSKFMGRTLEACNSYVGTCAYMSPE 224
HRDIK +NIL++ +VK+ADFGVS + RT+ ++VGT +M+PE
Sbjct: 543 IHRDIKAANILLSENGDVKVADFGVSAQLTRTISRRKTFVGTPFWMAPE 689
Score = 25.4 bits (54), Expect(2) = 3e-23
Identities = 10/16 (62%), Positives = 12/16 (74%)
Frame = +1
Query: 240 ADIWSLGLTLFELYVG 255
ADIWSLG+T E+ G
Sbjct: 724 ADIWSLGITAIEMAKG 771
>TC88402 similar to GP|20804521|dbj|BAB92215. putative CRK1 protein {Oryza
sativa (japonica cultivar-group)}, partial (46%)
Length = 2184
Score = 103 bits (257), Expect = 9e-23
Identities = 70/197 (35%), Positives = 99/197 (49%), Gaps = 3/197 (1%)
Frame = +1
Query: 62 LGHGNGGTVYKVRHKLTSIIYALKINHYDS-DPTTRRRALTEVNILRRATDCTNVVKYHG 120
LG G TVYK R I ALK +D+ DP + + E++ILRR D N++K G
Sbjct: 994 LGKGTYSTVYKARDVTNQKIVALKRVRFDNLDPESVKFMAREIHILRRL-DHPNIIKLEG 1170
Query: 121 SFEKPTG-DVCILMEYMDSGSLETALKTTGTFSESKLSTVARDILNGLTYLHARNIAHRD 179
T + ++ EYM+ A + FSE +L +L+GL + H+ + HRD
Sbjct: 1171 LITSETSRSLYLVFEYMEHDLTGLASNPSIKFSEPQLKCYMHQLLSGLDHCHSHGVLHRD 1350
Query: 180 IKPSNILVNIKNEVKIADFGVSKFMGRTLE-ACNSYVGTCAYMSPERFDPEVYGGNYNGF 238
IK SN+L++ +KIADFG++ L S V T Y PE + G N+ G
Sbjct: 1351 IKGSNLLIDNNGVLKIADFGLANVFDAHLNIPLTSRVVTLWYRPPEL----LLGANHYGV 1518
Query: 239 SADIWSLGLTLFELYVG 255
+ D+WS G L ELY G
Sbjct: 1519 AVDLWSTGCILRELYTG 1569
>TC86454 homologue to GP|4567091|gb|AAD23582.1| SNF-1-like serine/threonine
protein kinase {Glycine max}, partial (90%)
Length = 2194
Score = 100 bits (250), Expect = 6e-22
Identities = 74/224 (33%), Positives = 112/224 (49%), Gaps = 11/224 (4%)
Frame = +3
Query: 47 GGDNISA--GDFEKLSVLGHGNGGTVYKVRHKLTS------IIYALKINHYDSDPTTRRR 98
GG N++A +++ LG G+ G V H LT I+ KI + + + RR
Sbjct: 81 GGGNVNAFLRNYKMGKTLGIGSFGKVKIAEHVLTGHKVAIKILNRRKIKNMEMEEKVRR- 257
Query: 99 ALTEVNILRRATDCTNVVKYHGSFEKPTGDVCILMEYMDSGSLETALKTTGTFSESKLST 158
E+ ILR ++++ + E PT D+ ++MEY+ SG L + G E + +
Sbjct: 258 ---EIKILRLFMH-HHIIRLYEVVETPT-DIYVVMEYVKSGELFDYIVEKGRLQEDEARS 422
Query: 159 VARDILNGLTYLHARNIAHRDIKPSNILVNIKNEVKIADFGVSKFM--GRTLEACNSYVG 216
+ I++G+ Y H + HRD+KP N+L++ K VKIADFG+S M G L+
Sbjct: 423 FFQQIISGVEYCHRNMVVHRDLKPENLLLDSKWSVKIADFGLSNIMRDGHFLKT------ 584
Query: 217 TCAYMSPERFDPEVYGGN-YNGFSADIWSLGLTLFELYVGYFPF 259
+C SP PEV G Y G D+WS G+ L+ L G PF
Sbjct: 585 SCG--SPNYAAPEVISGKLYAGPEVDVWSCGVILYALLCGTLPF 710
>TC78370 similar to GP|5001828|gb|AAD37165.1| 3-phosphoinositide-dependent
protein kinase-1 {Arabidopsis thaliana}, partial (91%)
Length = 2137
Score = 99.4 bits (246), Expect = 2e-21
Identities = 81/298 (27%), Positives = 139/298 (46%), Gaps = 19/298 (6%)
Frame = +1
Query: 39 SATTASVAGGDNISAGDFEKLSVLGHGNGGTVYKVRHKLTSIIYALKINHYDSDPTTRRR 98
S + A A +N + DFE + G G+ V + + K T ++YA+KI D T+
Sbjct: 424 SKSFAFRAPHENYTIQDFELGKIYGVGSYSKVVRAKKKDTGVVYAMKI--MDKKFITKEN 597
Query: 99 ALTEVNILRRATDCTN---VVKYHGSFEKPTGDVCILMEYMDSGSLETALKTTGTFSESK 155
V + R D + +V+ + +F+ T + + +E + G L + ++ +
Sbjct: 598 KTAYVKLERIVLDQLDHPGIVRLYFTFQD-TFSLYMALESCEGGELFDQITRKSRLTQDE 774
Query: 156 LSTVARDILNGLTYLHARNIAHRDIKPSNILVNIKNEVKIADFGVSKFMGRTL------- 208
A ++++ L Y+H+ + HRDIKP N+L+ + +KIADFG K M +
Sbjct: 775 AQFYAAEVVDALEYIHSLGVIHRDIKPENLLLTTEGHIKIADFGSVKPMQDSQITVLPNA 954
Query: 209 ----EACNSYVGTCAYMSPERFDPEVYGGNYNGFSADIWSLGLTLFELYVGYFPFLQSGQ 264
+AC ++VGT AY+ PEV + F D+W+LG TL+++ G PF +
Sbjct: 955 ASDDKAC-TFVGTAAYV-----PPEVLNSSPATFGNDLWALGCTLYQMLSGTSPFKDAS- 1113
Query: 265 RPDWASLMCAICFSDPPSLPETASSEFRNFVECCLKKESGERWSA-----AQLLTHPF 317
+W L+ + P+ S E R+ ++ L + R A A L +HPF
Sbjct: 1114--EW--LIFQRIIAREIRFPDYFSDEARDLIDRLLDLDPSRRPGAGPDGYAILKSHPF 1275
>TC90642 similar to GP|16904222|gb|AAL30818.1 calcium/calmodulin-dependent
protein kinase CaMK1 {Nicotiana tabacum}, partial (50%)
Length = 933
Score = 99.0 bits (245), Expect = 2e-21
Identities = 71/223 (31%), Positives = 119/223 (52%), Gaps = 6/223 (2%)
Frame = +1
Query: 102 EVNILRRATDCTNVVKYHGSFEKPTGDVCILMEYMDSGSL-ETALKTTGTFSESKLSTVA 160
EV +L+ + N+VK++ +FE +V I+ME + G L + L G ++E +
Sbjct: 184 EVKMLKALSGHRNLVKFYDAFED-VNNVYIVMELCEGGELLDRILDRGGRYTEEDAKVIL 360
Query: 161 RDILNGLTYLHARNIAHRDIKPSNILVNIKNE---VKIADFGVSKFMGRTLEACNSYVGT 217
ILN + + H + + HRD+KP N L K+E +K+ DFG+S F+ R + N VG+
Sbjct: 361 LQILNVVAFCHLQGVVHRDLKPENFLFVSKDEDAVLKVIDFGLSDFV-RPEQRLNDIVGS 537
Query: 218 CAYMSPERFDPEVYGGNYNGFSADIWSLGLTLFELYVGYFPFLQSGQRPDW--ASLMCAI 275
Y++ PEV +Y+ AD+WS+G+ + L++ GQ ++ + L
Sbjct: 538 AYYVA-----PEVLHRSYS-VEADLWSVGVISYILFMWK*TVFGHGQNQEFFRSVLRANP 699
Query: 276 CFSDPPSLPETASSEFRNFVECCLKKESGERWSAAQLLTHPFL 318
F D P + S E ++FV+ L K+ +R +AAQ L+HP+L
Sbjct: 700 NFDDSPW--PSISPEAKDFVKRLLNKDHRKRMTAAQALSHPWL 822
>TC77566 similar to GP|310580|gb|AAA34017.1|| protein kinase 2 {Glycine
max}, partial (88%)
Length = 1502
Score = 96.3 bits (238), Expect = 1e-20
Identities = 76/271 (28%), Positives = 129/271 (47%), Gaps = 4/271 (1%)
Frame = +1
Query: 56 FEKLSVLGHGNGGTVYKVRHKLTSIIYALKINHYDSDPTTRRRALTEVNILRRATDCTNV 115
+E L LG GN G + K T + ALK + + E+ I R+ N+
Sbjct: 76 YEPLKELGSGNFGVARLAKDKNTGELVALK--YIERGKKIDENVQREI-INHRSLRHPNI 246
Query: 116 VKYHGSFEKPTGDVCILMEYMDSGSLETALKTTGTFSESKLSTVARDILNGLTYLHARNI 175
+++ P+ + I++EY G L + + G FSE + + +++G++Y H+ I
Sbjct: 247 IRFKEVLLTPS-HLAIVLEYASGGELFERICSAGRFSEDEARYFFQQLISGVSYCHSMEI 423
Query: 176 AHRDIKPSNILV--NIKNEVKIADFGVSKFMGRTLEACNSYVGTCAYMSPERFDPEVYGG 233
HRD+K N L+ N +KI DFG SK S VGT AY++PE +
Sbjct: 424 CHRDLKLENTLLDGNPSPRLKICDFGYSK-SAILHSQPKSTVGTPAYIAPEVLSRK---- 588
Query: 234 NYNGFSADIWSLGLTLFELYVGYFPFLQSGQRPDWASLMCAICFSDPPSLPE--TASSEF 291
Y+G AD+WS G+TL+ + VG +PF + ++ + I S+P+ S+E
Sbjct: 589 EYDGKIADVWSCGVTLYVMLVGAYPFEEPEDPRNFRKTIGRI-IGVQYSIPDYVRISAEC 765
Query: 292 RNFVECCLKKESGERWSAAQLLTHPFLCKDM 322
RN + + +R + ++ +P+ K+M
Sbjct: 766 RNLLSRIFVADPAKRVTIPEIKQNPWFLKNM 858
>TC77246 similar to PIR|C71408|C71408 probable protein kinase - Arabidopsis
thaliana, partial (65%)
Length = 1715
Score = 96.3 bits (238), Expect = 1e-20
Identities = 65/237 (27%), Positives = 110/237 (45%), Gaps = 17/237 (7%)
Frame = +3
Query: 51 ISAGDFEKLSVLGHGNGGTVYKVRHKLTSIIYALKINHYDS--DPTTRRRALTEVNILRR 108
I G ++ LG GN VY+ R + A+K+ D + R + E++ +RR
Sbjct: 72 IILGKYQLTKFLGRGNFAKVYQARSLIDGSTVAVKVIDKSKTVDASMEPRIVREIDAMRR 251
Query: 109 ATDCTNVVKYHGSFEKPTGDVCILMEYMDSGSLETALKTTGTFSESKLSTVARDILNGLT 168
+ N++K H T + +++++ G L L F E+ + +++ L
Sbjct: 252 LQNHPNILKIHEVLATKT-KIYLIVDFAGGGELFLKLARRRKFPEALARRYFQQLVSALC 428
Query: 169 YLHARNIAHRDIKPSNILVNIKNEVKIADFGVSKFMGRTLEA-CNSYVGTCAYMSPERFD 227
+ H IAHRD+KP N+L++ + +K++DFG+S + ++ GT AY +
Sbjct: 429 FCHRNGIAHRDLKPQNLLLDAEGNLKVSDFGLSALPEQLKNGLLHTACGTPAYTA----- 593
Query: 228 PEVYGG-NYNGFSADIWSLGLTLFELYVGYFPFLQSG-------------QRPDWAS 270
PE+ G Y+G AD WS G+ L+ L G+ PF S Q P+W S
Sbjct: 594 PEILGRIGYDGSKADAWSCGVILYVLLAGHLPFDDSNIPAMCKRITRRDYQFPEWIS 764
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.319 0.135 0.410
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,651,149
Number of Sequences: 36976
Number of extensions: 155502
Number of successful extensions: 1496
Number of sequences better than 10.0: 500
Number of HSP's better than 10.0 without gapping: 1268
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1298
length of query: 324
length of database: 9,014,727
effective HSP length: 96
effective length of query: 228
effective length of database: 5,465,031
effective search space: 1246027068
effective search space used: 1246027068
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 58 (26.9 bits)
Medicago: description of AC139706.8


