

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC139601.8 - phase: 0
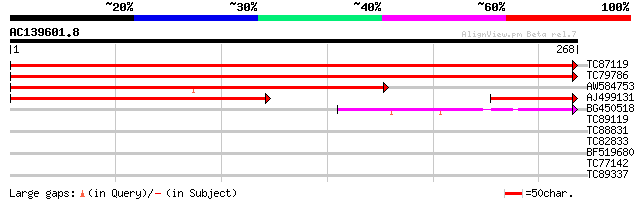
(268 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC87119 similar to GP|20197209|gb|AAM14973.1 expressed protein {... 543 e-155
TC79786 similar to GP|20197209|gb|AAM14973.1 expressed protein {... 440 e-124
AW584753 similar to GP|20197209|gb expressed protein {Arabidopsi... 281 2e-76
AJ499131 weakly similar to GP|22324441|db contains ESTs AU091908... 249 6e-67
BG450518 40 7e-04
TC89119 similar to PIR|T51958|T51958 hypothetical protein [impor... 29 1.7
TC88831 similar to GP|9758871|dbj|BAB09425.1 gb|AAD20092.1~gene_... 29 2.2
TC82833 similar to PIR|T48188|T48188 aldose reductase-like prote... 29 2.2
BF519680 weakly similar to GP|13160399|em aldose reductase {Digi... 28 2.8
TC77142 homologue to GP|9927482|emb|CAC04887.1 unnamed protein p... 28 4.8
TC89337 similar to GP|21592967|gb|AAM64916.1 putative GDSL-motif... 27 6.3
>TC87119 similar to GP|20197209|gb|AAM14973.1 expressed protein {Arabidopsis
thaliana}, complete
Length = 1424
Score = 543 bits (1399), Expect = e-155
Identities = 268/268 (100%), Positives = 268/268 (100%)
Frame = +1
Query: 1 MGHLYALDFDGVLCDTCGETAISALKAAKLRWPDLFGSVDSSTEDWIVEQMIKVRPVVET 60
MGHLYALDFDGVLCDTCGETAISALKAAKLRWPDLFGSVDSSTEDWIVEQMIKVRPVVET
Sbjct: 232 MGHLYALDFDGVLCDTCGETAISALKAAKLRWPDLFGSVDSSTEDWIVEQMIKVRPVVET 411
Query: 61 GYETLLLVRLLLETRVPSIRKSSVAEGLTVEGILEDWFKLKPIVMEEWNENRDDLIDLFG 120
GYETLLLVRLLLETRVPSIRKSSVAEGLTVEGILEDWFKLKPIVMEEWNENRDDLIDLFG
Sbjct: 412 GYETLLLVRLLLETRVPSIRKSSVAEGLTVEGILEDWFKLKPIVMEEWNENRDDLIDLFG 591
Query: 121 KVRDDWLENDFAGWIQGNRFYPGVADALRFASSKVYIVTTKQGRFADALLRELAGITIPP 180
KVRDDWLENDFAGWIQGNRFYPGVADALRFASSKVYIVTTKQGRFADALLRELAGITIPP
Sbjct: 592 KVRDDWLENDFAGWIQGNRFYPGVADALRFASSKVYIVTTKQGRFADALLRELAGITIPP 771
Query: 181 ERIYGLGTGPKVETLKKLQKMPEHQGLTLHFVEDRIAALKNVIKEPELDNWNLYLVNWGF 240
ERIYGLGTGPKVETLKKLQKMPEHQGLTLHFVEDRIAALKNVIKEPELDNWNLYLVNWGF
Sbjct: 772 ERIYGLGTGPKVETLKKLQKMPEHQGLTLHFVEDRIAALKNVIKEPELDNWNLYLVNWGF 951
Query: 241 NTQKERDEAAANPRIQLIDLSDFSSKLK 268
NTQKERDEAAANPRIQLIDLSDFSSKLK
Sbjct: 952 NTQKERDEAAANPRIQLIDLSDFSSKLK 1035
>TC79786 similar to GP|20197209|gb|AAM14973.1 expressed protein {Arabidopsis
thaliana}, complete
Length = 972
Score = 440 bits (1132), Expect = e-124
Identities = 213/268 (79%), Positives = 237/268 (87%)
Frame = +2
Query: 1 MGHLYALDFDGVLCDTCGETAISALKAAKLRWPDLFGSVDSSTEDWIVEQMIKVRPVVET 60
MG LYALDFDGV+CD+CGE+++SALKAAK+RWP LF VDS+ EDWIV+QM VRPVVET
Sbjct: 23 MGELYALDFDGVICDSCGESSLSALKAAKVRWPVLFDGVDSTIEDWIVDQMHTVRPVVET 202
Query: 61 GYETLLLVRLLLETRVPSIRKSSVAEGLTVEGILEDWFKLKPIVMEEWNENRDDLIDLFG 120
GYE LLLVRLLLETR PSIRKSSVAEGL VEGILE W LKPI+MEEW ENR+ LIDLFG
Sbjct: 203 GYENLLLVRLLLETRTPSIRKSSVAEGLAVEGILESWSTLKPIIMEEWGENREALIDLFG 382
Query: 121 KVRDDWLENDFAGWIQGNRFYPGVADALRFASSKVYIVTTKQGRFADALLRELAGITIPP 180
KVRD+WLE DFA WI NR YPGV+DAL+FASSKV+IVTTKQ RFADALLRELAG+TIP
Sbjct: 383 KVRDEWLEQDFAAWIGANRIYPGVSDALKFASSKVFIVTTKQSRFADALLRELAGVTIPS 562
Query: 181 ERIYGLGTGPKVETLKKLQKMPEHQGLTLHFVEDRIAALKNVIKEPELDNWNLYLVNWGF 240
ERIYGLGTGPKVE LK+LQK PEHQGLTLHFVEDR+A LKNVIKEPELD WNLYL NWG+
Sbjct: 563 ERIYGLGTGPKVEILKQLQKRPEHQGLTLHFVEDRLATLKNVIKEPELDKWNLYLGNWGY 742
Query: 241 NTQKERDEAAANPRIQLIDLSDFSSKLK 268
NT +E++EAAA PRI+++ LSDFS KLK
Sbjct: 743 NTAQEKEEAAAIPRIRVLQLSDFSKKLK 826
>AW584753 similar to GP|20197209|gb expressed protein {Arabidopsis thaliana},
partial (66%)
Length = 613
Score = 281 bits (719), Expect = 2e-76
Identities = 140/196 (71%), Positives = 158/196 (80%), Gaps = 17/196 (8%)
Frame = +3
Query: 1 MGHLYALDFDGVLCDTCGETAISALKAAKLRWPDLFGSVDSSTEDWIVEQMIKVRPVVET 60
MG LYALDFDGV+CD+CGE+++SALKAAK+RWP LF VDS+ EDWIV+QM VRPVVET
Sbjct: 24 MGELYALDFDGVICDSCGESSLSALKAAKVRWPVLFDGVDSTIEDWIVDQMHTVRPVVET 203
Query: 61 GYETLLLVRLLLETRVPSIRKSSVA-----------------EGLTVEGILEDWFKLKPI 103
GYE LLLVRLLLETR PSIRKSSV EGL VEGILE W LKPI
Sbjct: 204 GYENLLLVRLLLETRTPSIRKSSVGKIPIPHFLSLITMQPRLEGLAVEGILESWSTLKPI 383
Query: 104 VMEEWNENRDDLIDLFGKVRDDWLENDFAGWIQGNRFYPGVADALRFASSKVYIVTTKQG 163
+MEEW ENR+ LIDLFGKVRD+WLE+DFA WI NR YPGV+DAL+FASS+V+IVTTKQ
Sbjct: 384 IMEEWGENREALIDLFGKVRDEWLEHDFAAWIGANRIYPGVSDALKFASSEVFIVTTKQS 563
Query: 164 RFADALLRELAGITIP 179
RFADA+LRELAG+TIP
Sbjct: 564 RFADAILRELAGVTIP 611
>AJ499131 weakly similar to GP|22324441|db contains ESTs AU091908(E10074)
C19184(E10074)~similar to Arabidopsis thaliana
chromosome 2, partial (59%)
Length = 635
Score = 249 bits (637), Expect = 6e-67
Identities = 123/123 (100%), Positives = 123/123 (100%)
Frame = +1
Query: 1 MGHLYALDFDGVLCDTCGETAISALKAAKLRWPDLFGSVDSSTEDWIVEQMIKVRPVVET 60
MGHLYALDFDGVLCDTCGETAISALKAAKLRWPDLFGSVDSSTEDWIVEQMIKVRPVVET
Sbjct: 4 MGHLYALDFDGVLCDTCGETAISALKAAKLRWPDLFGSVDSSTEDWIVEQMIKVRPVVET 183
Query: 61 GYETLLLVRLLLETRVPSIRKSSVAEGLTVEGILEDWFKLKPIVMEEWNENRDDLIDLFG 120
GYETLLLVRLLLETRVPSIRKSSVAEGLTVEGILEDWFKLKPIVMEEWNENRDDLIDLFG
Sbjct: 184 GYETLLLVRLLLETRVPSIRKSSVAEGLTVEGILEDWFKLKPIVMEEWNENRDDLIDLFG 363
Query: 121 KVR 123
KVR
Sbjct: 364 KVR 372
Score = 88.6 bits (218), Expect = 2e-18
Identities = 41/41 (100%), Positives = 41/41 (100%)
Frame = +2
Query: 228 LDNWNLYLVNWGFNTQKERDEAAANPRIQLIDLSDFSSKLK 268
LDNWNLYLVNWGFNTQKERDEAAANPRIQLIDLSDFSSKLK
Sbjct: 368 LDNWNLYLVNWGFNTQKERDEAAANPRIQLIDLSDFSSKLK 490
>BG450518
Length = 673
Score = 40.4 bits (93), Expect = 7e-04
Identities = 34/118 (28%), Positives = 52/118 (43%), Gaps = 5/118 (4%)
Frame = +3
Query: 156 YIVTTKQGRFADALLRELAGITIP---PERIYGLGTGPKVETLKKLQKMP--EHQGLTLH 210
+ +T+K + L + AGI+IP P+ GL V L ++ P + G L+
Sbjct: 228 FYITSKLESAQISKLLQGAGISIPSDSPQLFTGLDPQQTVGVLTQISSRPVVKENGARLN 407
Query: 211 FVEDRIAALKNVIKEPELDNWNLYLVNWGFNTQKERDEAAANPRIQLIDLSDFSSKLK 268
F+ AL K LDN LYL W + + ER + P +QL++ LK
Sbjct: 408 FLSASSEALTAASK---LDNIRLYLAGW--SQEGERSKGGVGPDVQLLEEKQLXELLK 566
>TC89119 similar to PIR|T51958|T51958 hypothetical protein [imported] -
Picea mariana, partial (42%)
Length = 862
Score = 29.3 bits (64), Expect = 1.7
Identities = 17/55 (30%), Positives = 23/55 (40%), Gaps = 5/55 (9%)
Frame = +2
Query: 206 GLTLHFVEDRIAALKNVIKEPELDN-----WNLYLVNWGFNTQKERDEAAANPRI 255
G+T+ D ++ + I PE N WN W N + ER A PRI
Sbjct: 458 GITMSISSDLQSSSSSQISPPESVNQGLVQWNQIRQQWAGNKRSERQTVAREPRI 622
>TC88831 similar to GP|9758871|dbj|BAB09425.1
gb|AAD20092.1~gene_id:K24G6.5~similar to unknown protein
{Arabidopsis thaliana}, partial (7%)
Length = 880
Score = 28.9 bits (63), Expect = 2.2
Identities = 18/64 (28%), Positives = 32/64 (49%), Gaps = 2/64 (3%)
Frame = +2
Query: 165 FADALL--RELAGITIPPERIYGLGTGPKVETLKKLQKMPEHQGLTLHFVEDRIAALKNV 222
F DA+L + + TI R+ G+G G KV + Q + H+ F+E + ++
Sbjct: 92 FVDAVLPHKWITTTTITKTRLLGIGIGRKVIVSRSHQILTFHKSCGTKFLELKTFLSSSM 271
Query: 223 IKEP 226
+K+P
Sbjct: 272 MKQP 283
>TC82833 similar to PIR|T48188|T48188 aldose reductase-like protein -
Arabidopsis thaliana, partial (66%)
Length = 794
Score = 28.9 bits (63), Expect = 2.2
Identities = 12/23 (52%), Positives = 17/23 (73%)
Frame = +1
Query: 218 ALKNVIKEPELDNWNLYLVNWGF 240
AL N ++E +LD +LYLV+W F
Sbjct: 70 ALNNTLQELQLDYLDLYLVHWPF 138
>BF519680 weakly similar to GP|13160399|em aldose reductase {Digitalis
purpurea}, partial (43%)
Length = 569
Score = 28.5 bits (62), Expect = 2.8
Identities = 12/31 (38%), Positives = 20/31 (63%)
Frame = +3
Query: 213 EDRIAALKNVIKEPELDNWNLYLVNWGFNTQ 243
ED AL +++ +LD +LYL++W F T+
Sbjct: 408 EDVSKALARTLEDLQLDYIDLYLIHWPFRTK 500
>TC77142 homologue to GP|9927482|emb|CAC04887.1 unnamed protein product
{Medicago sativa}, complete
Length = 1350
Score = 27.7 bits (60), Expect = 4.8
Identities = 17/59 (28%), Positives = 29/59 (48%)
Frame = +3
Query: 210 HFVEDRIAALKNVIKEPELDNWNLYLVNWGFNTQKERDEAAANPRIQLIDLSDFSSKLK 268
H ED AL + + +LD +LYL++W + ++ E A + +D +D S K
Sbjct: 273 HHPEDVPKALDKTLNDLQLDYLDLYLIHWPVSMKRGTGEFKA----ENLDRADIPSTWK 437
>TC89337 similar to GP|21592967|gb|AAM64916.1 putative GDSL-motif
lipase/acylhydrolase {Arabidopsis thaliana}, partial
(65%)
Length = 881
Score = 27.3 bits (59), Expect = 6.3
Identities = 11/28 (39%), Positives = 18/28 (64%)
Frame = +3
Query: 216 IAALKNVIKEPELDNWNLYLVNWGFNTQ 243
IA LK+ IK + +W+ + +W +NTQ
Sbjct: 303 IAILKS*IKRRKASSWS*FCFSWNWNTQ 386
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.319 0.139 0.419
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,824,228
Number of Sequences: 36976
Number of extensions: 98410
Number of successful extensions: 385
Number of sequences better than 10.0: 22
Number of HSP's better than 10.0 without gapping: 381
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 384
length of query: 268
length of database: 9,014,727
effective HSP length: 94
effective length of query: 174
effective length of database: 5,538,983
effective search space: 963783042
effective search space used: 963783042
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 57 (26.6 bits)
Medicago: description of AC139601.8


