

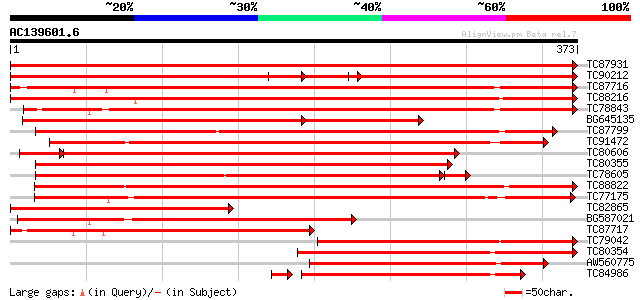
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC139601.6 - phase: 0
(373 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC87931 weakly similar to GP|5031283|gb|AAD38147.1| unknown {Pru... 754 0.0
TC90212 weakly similar to GP|5031283|gb|AAD38147.1| unknown {Pru... 325 e-159
TC87716 weakly similar to GP|5031283|gb|AAD38147.1| unknown {Pru... 478 e-135
TC88216 weakly similar to GP|5031283|gb|AAD38147.1| unknown {Pru... 401 e-112
TC78843 weakly similar to GP|8843897|dbj|BAA97423.1 1-aminocyclo... 384 e-107
BG645135 weakly similar to PIR|D84713|D84 probable dioxygenase [... 286 e-103
TC87799 weakly similar to PIR|C84713|C84713 probable dioxygenase... 365 e-101
TC91472 weakly similar to PIR|S59548|S59548 1-aminocyclopropane-... 312 2e-85
TC80606 similar to GP|5031283|gb|AAD38147.1| unknown {Prunus arm... 296 4e-85
TC80355 weakly similar to GP|5031283|gb|AAD38147.1| unknown {Pru... 296 7e-81
TC78605 similar to GP|5031283|gb|AAD38147.1| unknown {Prunus arm... 289 2e-80
TC88822 weakly similar to PIR|T06406|T06406 ripening protein E8 ... 286 1e-77
TC77175 weakly similar to PIR|C84713|C84713 probable dioxygenase... 284 3e-77
TC82865 weakly similar to PIR|D86201|D86201 protein F12K11.6 [im... 248 4e-66
BG587021 weakly similar to PIR|D86201|D8 protein F12K11.6 [impor... 244 4e-65
TC87717 weakly similar to PIR|D86201|D86201 protein F12K11.6 [im... 243 7e-65
TC79042 weakly similar to GP|8885557|dbj|BAA97487.1 leucoanthocy... 226 1e-59
TC80354 similar to GP|1916643|gb|AAC49826.1| desacetoxyvindoline... 220 6e-58
AW560775 similar to GP|5031283|gb| unknown {Prunus armeniaca}, p... 211 5e-55
TC84986 similar to GP|5031283|gb|AAD38147.1| unknown {Prunus arm... 197 1e-51
>TC87931 weakly similar to GP|5031283|gb|AAD38147.1| unknown {Prunus
armeniaca}, partial (59%)
Length = 1323
Score = 754 bits (1946), Expect = 0.0
Identities = 373/373 (100%), Positives = 373/373 (100%)
Frame = +1
Query: 1 MVVENSNQLKESSDFTYDREAEVKAFDDSKVGVKGLVDSGVSKIPRMFYTRKLEISESTA 60
MVVENSNQLKESSDFTYDREAEVKAFDDSKVGVKGLVDSGVSKIPRMFYTRKLEISESTA
Sbjct: 82 MVVENSNQLKESSDFTYDREAEVKAFDDSKVGVKGLVDSGVSKIPRMFYTRKLEISESTA 261
Query: 61 SDSKLSIPIVDLKDIHINPAQRVEVIDQIRSACHEWGFFQVINHEIPIIVLDEMIDGICR 120
SDSKLSIPIVDLKDIHINPAQRVEVIDQIRSACHEWGFFQVINHEIPIIVLDEMIDGICR
Sbjct: 262 SDSKLSIPIVDLKDIHINPAQRVEVIDQIRSACHEWGFFQVINHEIPIIVLDEMIDGICR 441
Query: 121 FHEQDADVRKEFYTRDLKKKVSYYSNVRLFNGQGANWRDTFGFAIAPDPFNPDEVPQICR 180
FHEQDADVRKEFYTRDLKKKVSYYSNVRLFNGQGANWRDTFGFAIAPDPFNPDEVPQICR
Sbjct: 442 FHEQDADVRKEFYTRDLKKKVSYYSNVRLFNGQGANWRDTFGFAIAPDPFNPDEVPQICR 621
Query: 181 DIVIEYSQKIKNLGFTIFELLSEALGLNPSYLKEFGCAEGLFTLGHFYPTCPEPELTMGS 240
DIVIEYSQKIKNLGFTIFELLSEALGLNPSYLKEFGCAEGLFTLGHFYPTCPEPELTMGS
Sbjct: 622 DIVIEYSQKIKNLGFTIFELLSEALGLNPSYLKEFGCAEGLFTLGHFYPTCPEPELTMGS 801
Query: 241 TAHTDSTFMTLLLQDQLGGLQVLHEDKWVNVPPVHGALVVNIGDLLQLISNDKFVSVLHR 300
TAHTDSTFMTLLLQDQLGGLQVLHEDKWVNVPPVHGALVVNIGDLLQLISNDKFVSVLHR
Sbjct: 802 TAHTDSTFMTLLLQDQLGGLQVLHEDKWVNVPPVHGALVVNIGDLLQLISNDKFVSVLHR 981
Query: 301 VLSQNIGPRISVASFFLNSRDPIEGTSKIYGPIKELISEENPAIYKDITIKDFLAHFHTE 360
VLSQNIGPRISVASFFLNSRDPIEGTSKIYGPIKELISEENPAIYKDITIKDFLAHFHTE
Sbjct: 982 VLSQNIGPRISVASFFLNSRDPIEGTSKIYGPIKELISEENPAIYKDITIKDFLAHFHTE 1161
Query: 361 GRNVKFLLEPFKL 373
GRNVKFLLEPFKL
Sbjct: 1162GRNVKFLLEPFKL 1200
>TC90212 weakly similar to GP|5031283|gb|AAD38147.1| unknown {Prunus
armeniaca}, partial (70%)
Length = 1335
Score = 325 bits (833), Expect(2) = e-159
Identities = 156/195 (80%), Positives = 174/195 (89%)
Frame = +1
Query: 1 MVVENSNQLKESSDFTYDREAEVKAFDDSKVGVKGLVDSGVSKIPRMFYTRKLEISESTA 60
MVV+N+NQL+E++D TYDR+AEVKAFDDS+ GV GLV+SGVSKIPR F+ KL+I E++
Sbjct: 43 MVVKNTNQLEEATDSTYDRKAEVKAFDDSRAGVNGLVESGVSKIPRFFHAGKLDIGENST 222
Query: 61 SDSKLSIPIVDLKDIHINPAQRVEVIDQIRSACHEWGFFQVINHEIPIIVLDEMIDGICR 120
DSKLS+PIVDLKDIH NPAQRV+VI QIRSACHEWGFFQVINHEIPI VLDEMIDGI
Sbjct: 223 CDSKLSVPIVDLKDIHNNPAQRVDVIHQIRSACHEWGFFQVINHEIPITVLDEMIDGIRS 402
Query: 121 FHEQDADVRKEFYTRDLKKKVSYYSNVRLFNGQGANWRDTFGFAIAPDPFNPDEVPQICR 180
FHEQDADVRKEFYTRDLKKKV YYSNV LF+GQ ANWRDTFGFA+AP PF P+EVP ICR
Sbjct: 403 FHEQDADVRKEFYTRDLKKKVMYYSNVHLFSGQAANWRDTFGFAVAPHPFKPEEVPPICR 582
Query: 181 DIVIEYSQKIKNLGF 195
DIVIEYSQKIK +GF
Sbjct: 583 DIVIEYSQKIKGVGF 627
Score = 253 bits (647), Expect(2) = e-159
Identities = 121/150 (80%), Positives = 134/150 (88%)
Frame = +3
Query: 224 LGHFYPTCPEPELTMGSTAHTDSTFMTLLLQDQLGGLQVLHEDKWVNVPPVHGALVVNIG 283
LGH YP CPEPELTMG+T HTDS FMTLLLQDQLGGLQVLH+DKWVNVPPVHGALVVN+G
Sbjct: 714 LGHCYPPCPEPELTMGTTKHTDSNFMTLLLQDQLGGLQVLHKDKWVNVPPVHGALVVNVG 893
Query: 284 DLLQLISNDKFVSVLHRVLSQNIGPRISVASFFLNSRDPIEGTSKIYGPIKELISEENPA 343
DLLQLI+ND FVSV HRVLS NIGPRISVASFF+NS DPIEG SK+YGPIKEL+SEENP
Sbjct: 894 DLLQLITNDNFVSVYHRVLSHNIGPRISVASFFVNSPDPIEGKSKVYGPIKELLSEENPP 1073
Query: 344 IYKDITIKDFLAHFHTEGRNVKFLLEPFKL 373
IYKDITI+DFLAH++ +G + L+PFKL
Sbjct: 1074IYKDITIEDFLAHYYAKGLDGNSSLQPFKL 1163
Score = 66.2 bits (160), Expect = 2e-11
Identities = 34/62 (54%), Positives = 39/62 (62%)
Frame = +2
Query: 171 NPDEVPQICRDIVIEYSQKIKNLGFTIFELLSEALGLNPSYLKEFGCAEGLFTLGHFYPT 230
NP + Q + + +K + LGFTIFELLSEALGLN SYLKE CAEGLF G T
Sbjct: 554 NPRKCHQYAEIL*LNIHRK*RELGFTIFELLSEALGLNSSYLKELNCAEGLFIFGSLLST 733
Query: 231 CP 232
P
Sbjct: 734 VP 739
>TC87716 weakly similar to GP|5031283|gb|AAD38147.1| unknown {Prunus
armeniaca}, partial (66%)
Length = 1297
Score = 478 bits (1230), Expect = e-135
Identities = 234/377 (62%), Positives = 297/377 (78%), Gaps = 4/377 (1%)
Frame = +3
Query: 1 MVVENSNQLKESSDFTYDREAEVKAFDDSKVGVKGLVDSGV--SKIPRMFYTRKLEISES 58
M +E SN S TYDR AEVKAF++SK GVKGL++SG+ +KIPRMF++ L ++
Sbjct: 15 MELEASN----GSSSTYDRTAEVKAFEESKTGVKGLLESGIWTTKIPRMFHSPNLNLNSD 182
Query: 59 TASD--SKLSIPIVDLKDIHINPAQRVEVIDQIRSACHEWGFFQVINHEIPIIVLDEMID 116
S+ SK S+PI+DL+DI+ +P EV+++IRSAC EWGFFQVINH IP+IVLDEMI
Sbjct: 183 HESEASSKFSVPIIDLQDINTDPCLHAEVLEKIRSACKEWGFFQVINHGIPVIVLDEMIS 362
Query: 117 GICRFHEQDADVRKEFYTRDLKKKVSYYSNVRLFNGQGANWRDTFGFAIAPDPFNPDEVP 176
GI RFHEQDAD RK FYTRD KKV Y+SN LF ANWRD+ F ++P+P +P+E+P
Sbjct: 363 GIRRFHEQDADARKPFYTRDKSKKVRYFSNGSLFTDPAANWRDSLSFFVSPNPPDPEEIP 542
Query: 177 QICRDIVIEYSQKIKNLGFTIFELLSEALGLNPSYLKEFGCAEGLFTLGHFYPTCPEPEL 236
Q+CRDIVIEYS+K++ LG TIFEL SEALGL+PSYLK+ G F L H+YP CPEPEL
Sbjct: 543 QVCRDIVIEYSKKVRALGLTIFELFSEALGLHPSYLKDLALDYGQFLLCHYYPPCPEPEL 722
Query: 237 TMGSTAHTDSTFMTLLLQDQLGGLQVLHEDKWVNVPPVHGALVVNIGDLLQLISNDKFVS 296
TMG++ HTD FMT+LLQDQ+GGLQVLH+++WV+VPP+HG+LVVNIGDLLQL++ND F S
Sbjct: 723 TMGTSKHTDIDFMTILLQDQIGGLQVLHQNQWVHVPPLHGSLVVNIGDLLQLVTNDMFTS 902
Query: 297 VLHRVLSQNIGPRISVASFFLNSRDPIEGTSKIYGPIKELISEENPAIYKDITIKDFLAH 356
V HRVLS++IGPR+S+ASFF NS +E +SK+ GPIKEL+SEENPAIY+D T+K+ LAH
Sbjct: 903 VYHRVLSKHIGPRVSIASFFANS---VEVSSKVLGPIKELLSEENPAIYRDTTVKEVLAH 1073
Query: 357 FHTEGRNVKFLLEPFKL 373
+ T+G + L PF+L
Sbjct: 1074YFTKGLDGNSSLHPFRL 1124
>TC88216 weakly similar to GP|5031283|gb|AAD38147.1| unknown {Prunus
armeniaca}, partial (68%)
Length = 1508
Score = 401 bits (1030), Expect = e-112
Identities = 185/376 (49%), Positives = 270/376 (71%), Gaps = 3/376 (0%)
Frame = +2
Query: 1 MVVENSNQLKESSDFTYDREAEVKAFDDSKVGVKGLVDSGVSKIPRMFYTRKL-EISEST 59
MV N ++K+ + YDR+ E+K D+SK GVKGLVD+G++K+P++F K+ E +
Sbjct: 158 MVNTNLEEIKQEHEHVYDRQKELKLLDESKEGVKGLVDAGLTKVPKIFIHDKIHEHNNKQ 337
Query: 60 ASDSKLSIPIVDLKDIHINPAQ--RVEVIDQIRSACHEWGFFQVINHEIPIIVLDEMIDG 117
S + LSIPI+D + N + R+E+I++++ A +WGFFQV+NH IP VLDEMIDG
Sbjct: 338 TSSTNLSIPIIDFGPLFTNTSSSSRLEIIEKVKHASEKWGFFQVVNHGIPSTVLDEMIDG 517
Query: 118 ICRFHEQDADVRKEFYTRDLKKKVSYYSNVRLFNGQGANWRDTFGFAIAPDPFNPDEVPQ 177
+ RFHEQD +++K+FY+RD+ K+ + +N L+ NWRD+ + P P +P ++P
Sbjct: 518 VVRFHEQDTEMKKKFYSRDITKRAYFNTNFDLYVTPAVNWRDSLSCVMGPQPLDPQDLPT 697
Query: 178 ICRDIVIEYSQKIKNLGFTIFELLSEALGLNPSYLKEFGCAEGLFTLGHFYPTCPEPELT 237
+CRDI ++YS + +G + ELLSEALGLN +YLK+ CAEGLF + H+YP CPEPELT
Sbjct: 698 VCRDITVKYSDYVNKVGMILLELLSEALGLNSNYLKDIDCAEGLFLISHYYPPCPEPELT 877
Query: 238 MGSTAHTDSTFMTLLLQDQLGGLQVLHEDKWVNVPPVHGALVVNIGDLLQLISNDKFVSV 297
G++AH+DS+F T+LLQDQLGGLQV H ++WV+V P+ GALV+N+GD++QLI+NDKF+SV
Sbjct: 878 FGTSAHSDSSFFTVLLQDQLGGLQVFHGNQWVDVTPIPGALVINLGDMMQLITNDKFLSV 1057
Query: 298 LHRVLSQNIGPRISVASFFLNSRDPIEGTSKIYGPIKELISEENPAIYKDITIKDFLAHF 357
HRVL+ IGPRISVA FF P SK+YGPI EL++ ENP +YK+ ++K ++H+
Sbjct: 1058KHRVLAPKIGPRISVACFFRQHLPP--ENSKLYGPITELLTPENPPVYKETSVKGLVSHY 1231
Query: 358 HTEGRNVKFLLEPFKL 373
+ +G + L+ F++
Sbjct: 1232YGKGLDGNSALDHFRI 1279
>TC78843 weakly similar to GP|8843897|dbj|BAA97423.1
1-aminocyclopropane-1-carboxylate oxidase {Arabidopsis
thaliana}, partial (70%)
Length = 1240
Score = 384 bits (985), Expect = e-107
Identities = 185/368 (50%), Positives = 270/368 (73%), Gaps = 4/368 (1%)
Frame = +2
Query: 10 KESSDFTYDREAEVKAFDDSKVGVKGLVDSGVSKIPRMFYTR--KLEISESTASDSKLSI 67
K + D T++ E KAFD++K GVKGLVD+GV KIP +F+ + K EI+ +T++ I
Sbjct: 47 KPNLDSTFN---ERKAFDETKAGVKGLVDAGVEKIPSLFHHQPDKYEIANNTSN----VI 205
Query: 68 PIVDLKDI-HINPAQRVEVIDQIRSACHEWGFFQVINHEIPIIVLDEMIDGICRFHEQDA 126
P++DL DI + +P+ ++++++ AC GFFQV+NH IP+ VL+E+ DG+ RF+EQD
Sbjct: 206 PVIDLDDIDNKDPSIHQGIVEKVKEACETLGFFQVVNHGIPLSVLEELNDGVKRFYEQDT 385
Query: 127 DVRKEFYTRDLKKKVSYYSNVRLFNGQGANWRDTFGFAIAP-DPFNPDEVPQICRDIVIE 185
+ +K FYTRD+++ Y SNV +++ NW+D+FG +AP D P+E P +CRDI++
Sbjct: 386 EAKKSFYTRDMQRSFIYNSNVDIYSSPALNWKDSFGCNLAPPDTLKPEEFPVVCRDILLR 565
Query: 186 YSQKIKNLGFTIFELLSEALGLNPSYLKEFGCAEGLFTLGHFYPTCPEPELTMGSTAHTD 245
Y + + NLG +FELLSEALGLNP++LK+ CAEGL L H+YP CPEPELT+G+T H+D
Sbjct: 566 YGKHMMNLGTLLFELLSEALGLNPNHLKDMDCAEGLIALCHYYPPCPEPELTVGTTKHSD 745
Query: 246 STFMTLLLQDQLGGLQVLHEDKWVNVPPVHGALVVNIGDLLQLISNDKFVSVLHRVLSQN 305
+ F+T+LLQD +GGLQVL++DKW+++ PV GAL+VN+GDLLQLI+ND+F SV HRV++
Sbjct: 746 NDFLTVLLQDHIGGLQVLYDDKWIDITPVPGALIVNVGDLLQLITNDRFKSVEHRVVANE 925
Query: 306 IGPRISVASFFLNSRDPIEGTSKIYGPIKELISEENPAIYKDITIKDFLAHFHTEGRNVK 365
+GPRISVA FF I +SK+YGPIKEL+SE+NP Y++ T+ D++A+F +G +
Sbjct: 926 VGPRISVACFFCTG---IRSSSKLYGPIKELLSEDNPPKYRETTVSDYVAYFEKKGLDGT 1096
Query: 366 FLLEPFKL 373
L +K+
Sbjct: 1097SALTHYKI 1120
>BG645135 weakly similar to PIR|D84713|D84 probable dioxygenase [imported] -
Arabidopsis thaliana, partial (44%)
Length = 812
Score = 286 bits (731), Expect(2) = e-103
Identities = 138/187 (73%), Positives = 163/187 (86%)
Frame = +1
Query: 9 LKESSDFTYDREAEVKAFDDSKVGVKGLVDSGVSKIPRMFYTRKLEISESTASDSKLSIP 68
L+ES++ TYDR+ EVK FDDSK GV+GLV+ GVSKIPR+F+T KL+I E++ASDSKLS+P
Sbjct: 1 LEESTNSTYDRKTEVKNFDDSKAGVRGLVEHGVSKIPRIFHTGKLDIGENSASDSKLSVP 180
Query: 69 IVDLKDIHINPAQRVEVIDQIRSACHEWGFFQVINHEIPIIVLDEMIDGICRFHEQDADV 128
IVDLKDIHI+ AQRVEVI+QI+SACH GFFQVINH+IPI VLDEMI GI FHEQD DV
Sbjct: 181 IVDLKDIHIDQAQRVEVIEQIQSACHVGGFFQVINHDIPIDVLDEMIHGIRSFHEQDVDV 360
Query: 129 RKEFYTRDLKKKVSYYSNVRLFNGQGANWRDTFGFAIAPDPFNPDEVPQICRDIVIEYSQ 188
RKEFYTRDLKKKV Y+SN LF+GQ ANWRDT GFA+AP PF P+E+P ICRD VI+YSQ
Sbjct: 361 RKEFYTRDLKKKVMYFSNGTLFSGQAANWRDTIGFAVAPHPFKPNELPSICRDSVIKYSQ 540
Query: 189 KIKNLGF 195
KI+++GF
Sbjct: 541 KIRDVGF 561
Score = 107 bits (267), Expect(2) = e-103
Identities = 52/81 (64%), Positives = 60/81 (73%), Gaps = 1/81 (1%)
Frame = +2
Query: 193 LGFTIFELLSEALGLNPSYLKEFGCAEGLFTLGHFYPTCPEPELTMGSTAHTDSTFMTLL 252
LGF IFELLSEALGL+ YLKE C EGLF GH+YP CPEPELTMG+ HTD++F+TLL
Sbjct: 554 LGFIIFELLSEALGLDRDYLKELNCCEGLFIQGHYYPACPEPELTMGTAKHTDTSFITLL 733
Query: 253 LQDQLGGL-QVLHEDKWVNVP 272
LQDQL + + DKW P
Sbjct: 734 LQDQLWWVFKFFMGDKWGQCP 796
>TC87799 weakly similar to PIR|C84713|C84713 probable dioxygenase [imported]
- Arabidopsis thaliana, partial (65%)
Length = 1398
Score = 365 bits (936), Expect = e-101
Identities = 171/343 (49%), Positives = 245/343 (70%)
Frame = +1
Query: 18 DREAEVKAFDDSKVGVKGLVDSGVSKIPRMFYTRKLEISESTASDSKLSIPIVDLKDIHI 77
+R E+KAFD++K+GVKGLVD+G++KIPR+FY + +++ +P++DL +I
Sbjct: 85 ERVKELKAFDETKLGVKGLVDAGITKIPRIFYQPRDSTKKASELGHTTIVPVIDLANIEK 264
Query: 78 NPAQRVEVIDQIRSACHEWGFFQVINHEIPIIVLDEMIDGICRFHEQDADVRKEFYTRDL 137
+P R V++ IR A +GFFQ++NH IP+ L+EM DG+ F EQD++V+KEFYTR+
Sbjct: 265 DPFARKRVVESIRDASETFGFFQIVNHGIPVSTLEEMKDGVKSFFEQDSEVKKEFYTRE- 441
Query: 138 KKKVSYYSNVRLFNGQGANWRDTFGFAIAPDPFNPDEVPQICRDIVIEYSQKIKNLGFTI 197
K +YSN+ L+ A W+D+F +AP P P+++P +CRDI++EY ++K +G +
Sbjct: 442 HKPFMHYSNIALYTAPAATWKDSFLCNMAPIPPKPEDLPVVCRDILLEYLSQVKKVGTLL 621
Query: 198 FELLSEALGLNPSYLKEFGCAEGLFTLGHFYPTCPEPELTMGSTAHTDSTFMTLLLQDQL 257
FELLSEALGLN +YL + GCAEG + GH+YP CPEPELT+G++ H D F+TLLLQD +
Sbjct: 622 FELLSEALGLNSTYLTDIGCAEGFYAFGHYYPPCPEPELTLGTSKHVDVVFLTLLLQDHV 801
Query: 258 GGLQVLHEDKWVNVPPVHGALVVNIGDLLQLISNDKFVSVLHRVLSQNIGPRISVASFFL 317
GGLQ LH D W++VPP+ AL+VNIGD LQLI+NDKF SV HRV + +GPR+S+ASFF
Sbjct: 802 GGLQYLHRDMWIDVPPMPEALIVNIGDFLQLITNDKFKSVHHRVPANLVGPRVSIASFFG 981
Query: 318 NSRDPIEGTSKIYGPIKELISEENPAIYKDITIKDFLAHFHTE 360
P T++ +GPIKEL+S+EN A YK+ + F+ + T+
Sbjct: 982 TLHHP---TTRTFGPIKELLSDENSAKYKETSFPKFMDSYITK 1101
>TC91472 weakly similar to PIR|S59548|S59548
1-aminocyclopropane-1-carboxylate oxidase homolog (clone
2A6) - Arabidopsis thaliana, partial (47%)
Length = 967
Score = 312 bits (799), Expect = 2e-85
Identities = 155/329 (47%), Positives = 217/329 (65%), Gaps = 1/329 (0%)
Frame = +1
Query: 27 DDSKVGVKGLVDSGVSKIPRMFYTRKLEISESTASDSKL-SIPIVDLKDIHINPAQRVEV 85
D+SK GVKGL+DSG++K+PR F K + + A+ S L +P++D +R+E+
Sbjct: 1 DESKTGVKGLLDSGITKLPRFFIHPKESLPKHDATSSCLFHVPVIDFTGYE--ECRRLEI 174
Query: 86 IDQIRSACHEWGFFQVINHEIPIIVLDEMIDGICRFHEQDADVRKEFYTRDLKKKVSYYS 145
+++I+ A WGFFQ++NH +P+ V+D+M+ I FHEQ ++V+KE+Y+RD K KV Y+S
Sbjct: 175 LNEIKEASETWGFFQMVNHCVPVCVMDDMLKAIKEFHEQPSEVKKEWYSRDHKVKVRYFS 354
Query: 146 NVRLFNGQGANWRDTFGFAIAPDPFNPDEVPQICRDIVIEYSQKIKNLGFTIFELLSEAL 205
N L + ANWRDT F P +P P +CR+ VI+Y + I L + EL+SEAL
Sbjct: 355 NGDLLVAKAANWRDTIIFDFHDGPLDPQAYPLVCREAVIKYMKHILKLKEILSELMSEAL 534
Query: 206 GLNPSYLKEFGCAEGLFTLGHFYPTCPEPELTMGSTAHTDSTFMTLLLQDQLGGLQVLHE 265
GL YL C + + H+YPTCPEPELT G+T H+D + +T+LLQD +GGLQVLH+
Sbjct: 535 GLKRDYLANIECMKSETVVCHYYPTCPEPELTFGATKHSDPSSLTILLQDTIGGLQVLHQ 714
Query: 266 DKWVNVPPVHGALVVNIGDLLQLISNDKFVSVLHRVLSQNIGPRISVASFFLNSRDPIEG 325
+ WV++ P+HGALV NIGD +QLISNDKF SV HRVL+ ++GPR SVA
Sbjct: 715 NHWVDINPIHGALVANIGDFMQLISNDKFKSVEHRVLAGSVGPRASVACHV------YPN 876
Query: 326 TSKIYGPIKELISEENPAIYKDITIKDFL 354
SK Y PI+E S+ENP YK+ I ++L
Sbjct: 877 ASKKYRPIEEFTSDENPPKYKETDITEYL 963
>TC80606 similar to GP|5031283|gb|AAD38147.1| unknown {Prunus armeniaca},
partial (58%)
Length = 890
Score = 296 bits (759), Expect(2) = 4e-85
Identities = 144/262 (54%), Positives = 196/262 (73%), Gaps = 1/262 (0%)
Frame = +2
Query: 36 LVDSGVSKIPRMFYTRKLEISESTASDS-KLSIPIVDLKDIHINPAQRVEVIDQIRSACH 94
L+ V+ IPRMF+ + S S++S+S KL IP +DL DIH +P +R V+++IR A
Sbjct: 104 LLMXSVTSIPRMFHHEFDKDSTSSSSNSHKLVIPSIDLVDIHQDPTRRKIVVEKIREASE 283
Query: 95 EWGFFQVINHEIPIIVLDEMIDGICRFHEQDADVRKEFYTRDLKKKVSYYSNVRLFNGQG 154
WGFFQ++NH I + VLDEM +G+ RF EQD++V++E YTRD K + Y SN L++
Sbjct: 284 TWGFFQIVNHGIEVSVLDEMKNGVVRFFEQDSEVKRELYTRDPVKPLVYNSNFDLYSSPA 463
Query: 155 ANWRDTFGFAIAPDPFNPDEVPQICRDIVIEYSQKIKNLGFTIFELLSEALGLNPSYLKE 214
A WRDTF +AP+ NP+++P +CRDI++EY++++ LG +FELLSEALGL+P+YL E
Sbjct: 464 ATWRDTFYCFMAPNSPNPEDLPSVCRDIMLEYTKQVMKLGNLLFELLSEALGLDPNYLNE 643
Query: 215 FGCAEGLFTLGHFYPTCPEPELTMGSTAHTDSTFMTLLLQDQLGGLQVLHEDKWVNVPPV 274
C EGL + H+YP+CPEPELT+G T HTD+ F+T+LLQD +GGLQVL E+ WV+V PV
Sbjct: 644 MRCNEGLALVCHYYPSCPEPELTLGITKHTDNDFITVLLQDHIGGLQVLRENSWVDVSPV 823
Query: 275 HGALVVNIGDLLQLISNDKFVS 296
GALV+NIGDLLQLI+NDKF S
Sbjct: 824 PGALVINIGDLLQLITNDKFKS 889
Score = 36.2 bits (82), Expect(2) = 4e-85
Identities = 15/30 (50%), Positives = 23/30 (76%)
Frame = +3
Query: 7 NQLKESSDFTYDREAEVKAFDDSKVGVKGL 36
N++ ++ YDR +E+KAFD++K GVKGL
Sbjct: 15 NEISMTTKHDYDRASELKAFDETKDGVKGL 104
>TC80355 weakly similar to GP|5031283|gb|AAD38147.1| unknown {Prunus
armeniaca}, partial (61%)
Length = 878
Score = 296 bits (759), Expect = 7e-81
Identities = 141/276 (51%), Positives = 198/276 (71%), Gaps = 2/276 (0%)
Frame = +2
Query: 18 DREAEVKAFDDSKVGVKGLVDSGVSKIPRMFYTRKLEISESTASDSKLSI-PIVDLKDIH 76
+R E+KAFDD+K GVKGLVD GV KIP +F+ + ++T S++ I PI+D +
Sbjct: 50 NRLHELKAFDDTKTGVKGLVDKGVVKIPTLFHHPLDKFGKATNSNNTQHIIPIIDFANFG 229
Query: 77 INPAQRVEVIDQIRSACHEWGFFQVINHEIPIIVLDEMIDGICRFHEQDADVRKEFYTRD 136
+P R E+ +IR AC WGFFQV+NH IP+ VL++M +G+ RF EQD +V+KE Y+RD
Sbjct: 230 NDPKTRHEITSKIREACETWGFFQVVNHGIPLNVLEDMRNGVIRFFEQDVEVKKELYSRD 409
Query: 137 LKKKVSYYSNVRLFNGQGANWRDTFGFAIAPDPFNPDEVPQICRDIVIEYSQKIKNLGFT 196
+ SY SN L++ NWRDTF ++AP+ P ++P +CRD ++EY + + LG T
Sbjct: 410 PMRPFSYNSNFNLYSSSALNWRDTFVCSLAPNAPKPQDLPLVCRDELLEYGEYVTKLGMT 589
Query: 197 IFELLSEALGLNPSYLKEFGCAEGLFTLGHFYPTCPEPELTMGSTAHTDSTFMTLL-LQD 255
+FELLSEALGL+P++LK+ GCA+G+ +L H+YP CPEPELTMG+T H+DS F+TLL D
Sbjct: 590 LFELLSEALGLHPNHLKDMGCAKGILSLSHYYPACPEPELTMGTTKHSDSCFLTLLPSDD 769
Query: 256 QLGGLQVLHEDKWVNVPPVHGALVVNIGDLLQLISN 291
+GGLQV H++KW+++PP+ A VN GDLLQL +N
Sbjct: 770 HIGGLQVFHQNKWIDIPPIPDAPGVNGGDLLQLGTN 877
>TC78605 similar to GP|5031283|gb|AAD38147.1| unknown {Prunus armeniaca},
partial (58%)
Length = 961
Score = 289 bits (739), Expect(2) = 2e-80
Identities = 135/270 (50%), Positives = 194/270 (71%), Gaps = 1/270 (0%)
Frame = +3
Query: 18 DREAEVKAFDDSKVGVKGLVDSGVSKIPRMFYTRKLEISESTASDSKLSIPIVDLKDIHI 77
+R E+KAFD++K+GV+GLVD+G++K+PR+FY +++ S +IP++DL +I
Sbjct: 90 ERVKELKAFDETKLGVRGLVDAGITKVPRIFYQPPDSTKKASESGDTTTIPVIDLANILE 269
Query: 78 NPAQRVEVIDQIRSACHEWGFFQVINHEIPIIVLDEMIDGICRFHEQDADVRKEFYTRDL 137
+P R V++ +R A +GFFQ++NH IP+ L+EM DG+ F EQD++V+K++YTR+
Sbjct: 270 DPCARKRVVESVRDASEIFGFFQIVNHGIPVSTLNEMKDGVVSFFEQDSEVKKKYYTRER 449
Query: 138 KKKVSYYSNVRL-FNGQGANWRDTFGFAIAPDPFNPDEVPQICRDIVIEYSQKIKNLGFT 196
K V Y SN L + W+DTF AP+P P+++P +CR+I++EY + +G
Sbjct: 450 KPFV-YNSNYNLLYTSDPITWKDTFLCNRAPNPPKPEDLPAVCRNILLEYLNHVMKVGTL 626
Query: 197 IFELLSEALGLNPSYLKEFGCAEGLFTLGHFYPTCPEPELTMGSTAHTDSTFMTLLLQDQ 256
+FELLSEALGLNP+YL + GCAEGL GH+YP+CPEPELT+G+ H D F+T+LLQD
Sbjct: 627 VFELLSEALGLNPTYLIDIGCAEGLSAFGHYYPSCPEPELTIGTVKHADIDFITVLLQDH 806
Query: 257 LGGLQVLHEDKWVNVPPVHGALVVNIGDLL 286
+GGLQVLH+D WV+VPP+ ALVVNIGD L
Sbjct: 807 IGGLQVLHKDMWVDVPPIPEALVVNIGDFL 896
Score = 28.1 bits (61), Expect(2) = 2e-80
Identities = 13/17 (76%), Positives = 13/17 (76%)
Frame = +1
Query: 287 QLISNDKFVSVLHRVLS 303
Q ISNDKF S HRVLS
Sbjct: 898 QFISNDKFKSAQHRVLS 948
>TC88822 weakly similar to PIR|T06406|T06406 ripening protein E8 homolog -
tomato, partial (31%)
Length = 1112
Score = 286 bits (731), Expect = 1e-77
Identities = 151/359 (42%), Positives = 222/359 (61%), Gaps = 2/359 (0%)
Frame = +3
Query: 17 YDREAEVKAFDDSKVGVKGLVDSGVSKIPRMFYTRKLEISE-STASDSKLSIPIVDLKDI 75
YDR VK FD+SK+GVKGL DSG+ IP++F + +S+ + S SIP++DL ++
Sbjct: 48 YDRVKAVKEFDESKMGVKGLSDSGIKTIPQIFIHPQQTLSDIKSTSKQTTSIPLIDLSNV 227
Query: 76 HINPAQRVEVIDQIRSACHEWGFFQVINHEIPIIVLDEMIDGICRFHEQDADVRKEFYTR 135
+P R +I QI+ A WGFFQ+INH + + LD I+ I FH Q + + ++Y R
Sbjct: 228 -ASPNHRPNIIKQIKEAAKTWGFFQIINHNVNVSSLDNTINAIESFHNQPHETKSKYYKR 404
Query: 136 DLKKKVSYYSNVRLFNGQGANWRDTFGFAIAPDPFNPDEVPQICRDIVIEYSQKIKNLGF 195
D + V Y SN L+ A+W D+ +AP+ +E+P+ICR ++E+ +
Sbjct: 405 DEGRGVMYASNNDLYRTNAASWHDSLQAWMAPEAPKAEELPEICRKEMVEWDFHAAGVAE 584
Query: 196 TIFELLSEALGLNPSYLKEFGCAEGLFTLGHFYPTCPEPELTMGSTAHTDSTFMTLLLQD 255
+FELLSE LGL LKE E +GH YP CP+P+LTMG T HTD +TLLLQ+
Sbjct: 585 VVFELLSEGLGLERGKLKELSFCETRVIVGHCYPYCPQPDLTMGITPHTDPCAITLLLQN 764
Query: 256 QLGGLQVLHEDKWVNVPPVHGALVVNIGDLLQLISNDKFVSVLHRVL-SQNIGPRISVAS 314
Q+ GLQV +E +WV+V PV G ++VNIGD LQ+ISN ++ SV HRVL + N RIS+
Sbjct: 765 QVPGLQVKYEGEWVDVNPVQGGIIVNIGDFLQIISNGEYSSVQHRVLANSNKKSRISIVM 944
Query: 315 FFLNSRDPIEGTSKIYGPIKELISEENPAIYKDITIKDFLAHFHTEGRNVKFLLEPFKL 373
F S+ EG +GP+ EL+S E P +Y+D T ++F+ +F+++G + K ++ K+
Sbjct: 945 FLNLSKWRGEG---YHGPLPELLSPEKPPVYRDFTTQEFIENFYSKGIDAKSFVDKGKI 1112
>TC77175 weakly similar to PIR|C84713|C84713 probable dioxygenase [imported]
- Arabidopsis thaliana, partial (40%)
Length = 1425
Score = 284 bits (727), Expect = 3e-77
Identities = 151/359 (42%), Positives = 222/359 (61%), Gaps = 3/359 (0%)
Frame = +3
Query: 17 YDREAEVKAFDDSKVGVKGLVDSGVSKIPRMFYTRKLEISESTASDS--KLSIPIVDLKD 74
YDR VK FD++K GVKGL+DSG+ IP F +S+ T + IP +DL
Sbjct: 153 YDRIKAVKEFDETKSGVKGLIDSGIKTIPSFFIHPPEILSDLTPRSDFPQPEIPTIDLSA 332
Query: 75 IHINPAQRVEVIDQIRSACHEWGFFQVINHEIPIIVLDEMIDGICRFHEQDADVRKEFYT 134
+H + R V++Q+RSA GFFQVINH + ++ +I + +FHEQ AD RK+ Y
Sbjct: 333 VHHS---RAAVVEQLRSAASTVGFFQVINHGVAPELMRSVIGAMKKFHEQPADERKKVYC 503
Query: 135 RDLKKKVSYYSNVRLFNGQGANWRDTFGFAIAPDPFNPDEVPQICRDIVIEYSQKIKNLG 194
R++ VSY SNV LF + A+WRDT + P P E+P++CR V+E+ +++ +G
Sbjct: 504 REMGTGVSYISNVDLFASKAASWRDTLQIKMGPVPAEEKEIPEVCRKEVMEWDKEVVRVG 683
Query: 195 FTIFELLSEALGLNPSYLKEFGCAEGLFTLGHFYPTCPEPELTMGSTAHTDSTFMTLLLQ 254
+ LLSE LGL L E G ++G +GH+YP CP+P LT+G +H D +T+LLQ
Sbjct: 684 DILLGLLSEGLGLGEERLTELGLSQGRVMVGHYYPFCPQPNLTVGLNSHADPGALTVLLQ 863
Query: 255 DQLGGLQVLHEDKWVNVPPVHGALVVNIGDLLQLISNDKFVSVLHRVLSQ-NIGPRISVA 313
D +GGLQV + W+NV P+ GALV+NIGDLLQ+ISN+++ S HRVL+ + PR+S+A
Sbjct: 864 DHIGGLQVRTQHGWINVKPLDGALVINIGDLLQIISNEEYKSADHRVLANPSNEPRVSIA 1043
Query: 314 SFFLNSRDPIEGTSKIYGPIKELISEENPAIYKDITIKDFLAHFHTEGRNVKFLLEPFK 372
FLN + K++GP+ EL S + P++Y+D T+ +F+ F + + K L F+
Sbjct: 1044-VFLNPGN----REKLFGPLPELTSADKPSLYRDFTLNEFMTRFFKKELDGKSLTNFFR 1205
>TC82865 weakly similar to PIR|D86201|D86201 protein F12K11.6 [imported] -
Arabidopsis thaliana, partial (2%)
Length = 547
Score = 248 bits (632), Expect = 4e-66
Identities = 120/147 (81%), Positives = 138/147 (93%)
Frame = +2
Query: 1 MVVENSNQLKESSDFTYDREAEVKAFDDSKVGVKGLVDSGVSKIPRMFYTRKLEISESTA 60
MVV+N+NQL+E+++ TYDREAEVKAFDDSKVGVKGLV+SGVSKIPRMF+T K+EISE +A
Sbjct: 107 MVVKNTNQLEETTNSTYDREAEVKAFDDSKVGVKGLVESGVSKIPRMFHTGKMEISEDSA 286
Query: 61 SDSKLSIPIVDLKDIHINPAQRVEVIDQIRSACHEWGFFQVINHEIPIIVLDEMIDGICR 120
S+SKLSIPI+DLKDIH NPA+RVEVI QI+SACHEWGFFQ+INHEIPI VLDEMIDGI +
Sbjct: 287 SESKLSIPIIDLKDIHNNPARRVEVIGQIQSACHEWGFFQIINHEIPINVLDEMIDGISK 466
Query: 121 FHEQDADVRKEFYTRDLKKKVSYYSNV 147
FHEQDA+VRK+FYTRDL KKV YYSNV
Sbjct: 467 FHEQDANVRKDFYTRDLNKKVMYYSNV 547
>BG587021 weakly similar to PIR|D86201|D8 protein F12K11.6 [imported] -
Arabidopsis thaliana, partial (7%)
Length = 663
Score = 244 bits (623), Expect = 4e-65
Identities = 122/225 (54%), Positives = 161/225 (71%), Gaps = 2/225 (0%)
Frame = +2
Query: 6 SNQLKESSDFTYDREAEVKAFDDSKVGVKGLVDSGVSKIPRMFYTR--KLEISESTASDS 63
S + F+ ++A+V+AFDDSK GVKG++DSG++KIP +F+ K + T S
Sbjct: 2 SKTMASLESFSDLKKAQVQAFDDSKTGVKGILDSGITKIPSIFHVNLEKSTQNSPTHESS 181
Query: 64 KLSIPIVDLKDIHINPAQRVEVIDQIRSACHEWGFFQVINHEIPIIVLDEMIDGICRFHE 123
+IPI+DL+ I +RVE++DQI++A WGFFQVINH IP+ VLDEMI+GICRFHE
Sbjct: 182 FTTIPIIDLQHI-----KRVELVDQIKNASKNWGFFQVINHGIPLHVLDEMINGICRFHE 346
Query: 124 QDADVRKEFYTRDLKKKVSYYSNVRLFNGQGANWRDTFGFAIAPDPFNPDEVPQICRDIV 183
QDA+ +K FY+RD KKV Y+SN +LF ANWRDT FA P+ NP+E+P +CRDIV
Sbjct: 347 QDAEAKKPFYSRDKDKKVRYFSNGKLFRDMAANWRDTIAFAANPNLPNPEELPAVCRDIV 526
Query: 184 IEYSQKIKNLGFTIFELLSEALGLNPSYLKEFGCAEGLFTLGHFY 228
EYS+++K LG TIFELLSE LGLN SYLK CA+ L+ +G +Y
Sbjct: 527 AEYSKQVKALGVTIFELLSEVLGLNLSYLK*MECAKALYIMGQYY 661
>TC87717 weakly similar to PIR|D86201|D86201 protein F12K11.6 [imported] -
Arabidopsis thaliana, partial (6%)
Length = 653
Score = 243 bits (621), Expect = 7e-65
Identities = 125/209 (59%), Positives = 151/209 (71%), Gaps = 9/209 (4%)
Frame = +1
Query: 1 MVVENSNQLKESSDFTYDREAEVKAFDDSKVGVKGLVDSG--VSKIPRMFYTRKLEISES 58
M +E SN S+ TY+R AEVKAFD+SK GVKGLV+SG V+KIPRMF+ K + +
Sbjct: 31 MELEASNS---STTSTYNRIAEVKAFDESKAGVKGLVESGTCVTKIPRMFHFPKSSLKNN 201
Query: 59 TA-------SDSKLSIPIVDLKDIHINPAQRVEVIDQIRSACHEWGFFQVINHEIPIIVL 111
T S SKL +PI+DL+D++ NP VEV+D+IRSAC EWGFFQVINH IP+ VL
Sbjct: 202 THETILQIDSSSKLCVPIIDLQDMNTNPCLHVEVVDKIRSACKEWGFFQVINHGIPVSVL 381
Query: 112 DEMIDGICRFHEQDADVRKEFYTRDLKKKVSYYSNVRLFNGQGANWRDTFGFAIAPDPFN 171
DEM I RFHEQ+ D RK FYTRD KKV Y+SN LF ANWRDT F +PDP N
Sbjct: 382 DEMTSAIRRFHEQEVDARKPFYTRDTSKKVRYFSNGTLFRDPAANWRDTIAFFTSPDPPN 561
Query: 172 PDEVPQICRDIVIEYSQKIKNLGFTIFEL 200
P+E+PQ+CRDIVIEYS+K++ LG TIFEL
Sbjct: 562 PEEIPQVCRDIVIEYSKKVRALGLTIFEL 648
>TC79042 weakly similar to GP|8885557|dbj|BAA97487.1 leucoanthocyanidin
dioxygenase-like protein {Arabidopsis thaliana}, partial
(39%)
Length = 1665
Score = 226 bits (576), Expect = 1e-59
Identities = 108/171 (63%), Positives = 136/171 (79%)
Frame = +1
Query: 203 EALGLNPSYLKEFGCAEGLFTLGHFYPTCPEPELTMGSTAHTDSTFMTLLLQDQLGGLQV 262
EALGL+PSYL + G F L H+YP CPEPELTMG++ HTD FMT+LLQDQ+GG QV
Sbjct: 1 EALGLHPSYLTKLLSTYGQFLLCHYYPACPEPELTMGTSKHTDIDFMTILLQDQIGGFQV 180
Query: 263 LHEDKWVNVPPVHGALVVNIGDLLQLISNDKFVSVLHRVLSQNIGPRISVASFFLNSRDP 322
LH+++WV+VPP+HG+LVVN+GDL+QLI+ND F SV HRVLS+NIGPRIS+ASFF+N
Sbjct: 181 LHQNQWVDVPPLHGSLVVNVGDLMQLITNDMFTSVYHRVLSKNIGPRISIASFFVNPSSE 360
Query: 323 IEGTSKIYGPIKELISEENPAIYKDITIKDFLAHFHTEGRNVKFLLEPFKL 373
E TSK+ PIKEL+SEENP IY+D T+K+ LAH+ T+G + L PF+L
Sbjct: 361 -EVTSKVVAPIKELLSEENPPIYRDTTLKEVLAHYFTKGLDGNSSLHPFRL 510
>TC80354 similar to GP|1916643|gb|AAC49826.1| desacetoxyvindoline
4-hydroxylase {Catharanthus roseus}, partial (32%)
Length = 738
Score = 220 bits (561), Expect = 6e-58
Identities = 103/185 (55%), Positives = 145/185 (77%), Gaps = 1/185 (0%)
Frame = +3
Query: 190 IKNLGFTIFELLSEALGLNPSYLKEFGCAEGLFTLGHFYPTCPEPELTMGSTAHTDSTFM 249
++ G T+FELLSEALGL+P++LK+ GCA+G+ +L H+YP CPEPELTMG+T H+DS F+
Sbjct: 12 VRXXGMTLFELLSEALGLHPNHLKDMGCAKGILSLSHYYPACPEPELTMGTTKHSDSCFL 191
Query: 250 TLLLQ-DQLGGLQVLHEDKWVNVPPVHGALVVNIGDLLQLISNDKFVSVLHRVLSQNIGP 308
TLLLQ D +GGLQV H++KW+++PP+ ALVVN+GDLLQL++NDKF SV H+VL+ IGP
Sbjct: 192 TLLLQDDHIGGLQVFHQNKWIDIPPIPDALVVNVGDLLQLVTNDKFKSV*HKVLANIIGP 371
Query: 309 RISVASFFLNSRDPIEGTSKIYGPIKELISEENPAIYKDITIKDFLAHFHTEGRNVKFLL 368
R+SVA FF + S YGPIKE +SE+NP YK+ITI +++A+++T+G + L
Sbjct: 372 RVSVACFF---NADLNSFSNQYGPIKEFLSEDNPPKYKEITISEYVAYYNTKGIDGISAL 542
Query: 369 EPFKL 373
+ F++
Sbjct: 543 QHFRV 557
>AW560775 similar to GP|5031283|gb| unknown {Prunus armeniaca}, partial (41%)
Length = 464
Score = 211 bits (536), Expect = 5e-55
Identities = 97/157 (61%), Positives = 127/157 (80%)
Frame = +3
Query: 198 FELLSEALGLNPSYLKEFGCAEGLFTLGHFYPTCPEPELTMGSTAHTDSTFMTLLLQDQL 257
FEL SEAL LNP++LK+ CAE L LGH+YP CPEPELT+G+T H+D F+T+LLQD +
Sbjct: 3 FELCSEALSLNPNHLKDIDCAERLLALGHYYPPCPEPELTVGTTKHSDYDFLTVLLQDHI 182
Query: 258 GGLQVLHEDKWVNVPPVHGALVVNIGDLLQLISNDKFVSVLHRVLSQNIGPRISVASFFL 317
GGLQVL++DKW+++ PV GAL+VN+GDLLQLI+NDKF SV HRVL+ N+GPRISVA FF
Sbjct: 183 GGLQVLYQDKWIDITPVPGALIVNVGDLLQLITNDKFKSVEHRVLANNVGPRISVACFFC 362
Query: 318 NSRDPIEGTSKIYGPIKELISEENPAIYKDITIKDFL 354
SK+YGPIKEL+SE+NP+IY++ ++ D++
Sbjct: 363 TGH---MSFSKLYGPIKELLSEDNPSIYRETSVADYV 464
>TC84986 similar to GP|5031283|gb|AAD38147.1| unknown {Prunus armeniaca},
partial (42%)
Length = 530
Score = 197 bits (500), Expect(2) = 1e-51
Identities = 96/148 (64%), Positives = 120/148 (80%), Gaps = 1/148 (0%)
Frame = +3
Query: 193 LGFTIFELLSEALGLNPSYLKEFGCAEGLFTLGHFYPTCPEPELTMGSTAHTDSTFMTLL 252
LG +FELLSEALGL+P++LK+ C EG +GH+YP CPEP+LTMG+T H+D +F+T+L
Sbjct: 93 LGRVLFELLSEALGLHPNHLKDLDCCEGNMIIGHYYPACPEPDLTMGTTKHSDVSFLTVL 272
Query: 253 LQDQL-GGLQVLHEDKWVNVPPVHGALVVNIGDLLQLISNDKFVSVLHRVLSQNIGPRIS 311
LQD++ GGLQVLH+DKW++VPPV GALVVNIGDLLQLI+NDKF SV HRVL+ IGPRIS
Sbjct: 273 LQDRIIGGLQVLHQDKWIDVPPVPGALVVNIGDLLQLITNDKFKSVEHRVLANGIGPRIS 452
Query: 312 VASFFLNSRDPIEGTSKIYGPIKELISE 339
VA FF + + K+YGPI EL+SE
Sbjct: 453 VACFF---KAGLRAHKKLYGPITELLSE 527
Score = 24.3 bits (51), Expect(2) = 1e-51
Identities = 7/14 (50%), Positives = 13/14 (92%)
Frame = +2
Query: 173 DEVPQICRDIVIEY 186
+++P +CRDI++EY
Sbjct: 32 EDLPVVCRDILMEY 73
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.321 0.140 0.412
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,077,893
Number of Sequences: 36976
Number of extensions: 153084
Number of successful extensions: 1003
Number of sequences better than 10.0: 174
Number of HSP's better than 10.0 without gapping: 811
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 819
length of query: 373
length of database: 9,014,727
effective HSP length: 98
effective length of query: 275
effective length of database: 5,391,079
effective search space: 1482546725
effective search space used: 1482546725
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 59 (27.3 bits)
Medicago: description of AC139601.6


