

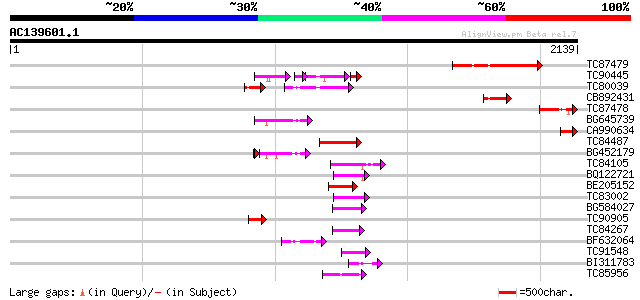
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC139601.1 - phase: 0 /pseudo
(2139 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC87479 similar to PIR|G84897|G84897 hypothetical protein At2g46... 446 e-125
TC90445 homologue to PIR|C84507|C84507 hypothetical protein At2g... 107 3e-53
TC80039 homologue to GP|6437558|gb|AAF08585.1| putative ATPase (... 185 2e-46
CB892431 similar to GP|10039641|gb| Orf122 {Chlorobium tepidum},... 158 2e-38
TC87478 homologue to PIR|G84897|G84897 hypothetical protein At2g... 142 1e-33
BG645739 similar to PIR|T45808|T4 helicase-like protein - Arabid... 136 8e-32
CA990634 homologue to PIR|G84897|G84 hypothetical protein At2g46... 135 2e-31
TC84487 similar to PIR|T52301|T52301 GYMNOS/PICKLE protein [impo... 132 1e-30
BG452179 homologue to GP|15528650|d putative DNA-dependent ATPas... 115 2e-30
TC84105 homologue to PIR|C84507|C84507 hypothetical protein At2g... 125 2e-28
BQ122721 similar to GP|8843733|dbj SWI2/SNF2-like protein {Arabi... 107 5e-23
BE205152 similar to GP|19347965|gb putative helicase {Arabidopsi... 107 6e-23
TC83002 similar to GP|14532630|gb|AAK64043.1 putative DNA-bindin... 84 6e-16
BG584027 similar to GP|10178052|db helicase-like transcription f... 74 6e-13
TC90905 similar to PIR|H84432|H84432 probable helicase [imported... 70 7e-12
TC84267 weakly similar to PIR|S41478|S41478 DNA repair protein r... 68 4e-11
BF632064 similar to PIR|H84432|H84 probable helicase [imported] ... 67 1e-10
TC91548 similar to PIR|D86185|D86185 hypothetical protein [impor... 64 5e-10
BI311783 homologue to PIR|T47587|T4 TATA box binding protein (TB... 55 4e-07
TC85956 homologue to GP|8885593|dbj|BAA97523.1 1 4-benzoquinone ... 54 5e-07
>TC87479 similar to PIR|G84897|G84897 hypothetical protein At2g46020
[imported] - Arabidopsis thaliana, partial (14%)
Length = 1031
Score = 446 bits (1148), Expect = e-125
Identities = 237/343 (69%), Positives = 274/343 (79%), Gaps = 1/343 (0%)
Frame = +3
Query: 1669 DELEEGEIAVSGESHMYHQQSGSWIHDRDEGEEEQVLQKPKIKRKRSLRVRPRHTMEKPE 1728
DELEEGEIAVS +SHM HQQSGSWIHDRDEGE+EQVLQKP+IKRKRS+RVRPRH EKPE
Sbjct: 12 DELEEGEIAVSFDSHMEHQQSGSWIHDRDEGEDEQVLQKPRIKRKRSIRVRPRHATEKPE 191
Query: 1729 DKSGSEMASLQRGQSFLLPDKKYPLQSRINQESKTFGDSSSNKHDKNEPILKNKRNLPAR 1788
DKSGSE Q+ D+KY Q R + ESK+ +S+++++++N I KNKR LP+R
Sbjct: 192 DKSGSETIPRLSVQA----DRKYQAQLRADLESKSHVESNASRNEQNSSI-KNKRTLPSR 356
Query: 1789 KVANASKLHVSPKSSRLNCTSAPSEDNDEHSRERLKGKPNNLRGSSAHVTNMTEIIQRRC 1848
+VAN SKLH SPK +RL SAPSED EHSRE +GKP N GSSAH + MTEIIQRRC
Sbjct: 357 RVANTSKLHSSPKPTRL---SAPSEDGGEHSRESWEGKPINSSGSSAHGSRMTEIIQRRC 527
Query: 1849 KSVISKLQRRIDKEGHQIVPLLTDLWKRIENSGFAGGSGNNLLDLRKIDQRINRLEYSGV 1908
K+VISKLQRRIDKEGHQIVPLLTDLWKRIENSG++GGSGNNLLDLRKIDQRI++LEY+G
Sbjct: 528 KNVISKLQRRIDKEGHQIVPLLTDLWKRIENSGYSGGSGNNLLDLRKIDQRIDKLEYTGA 707
Query: 1909 MEFVFDVQFMLKSAMQFYGYSYEVRTEARKVHDLFFDILKTTFSDIDFGEAKSALSFTSQ 1968
+ VFDVQFMLKSAMQ+YG+S EVRTEARKVH+LFFDILK F D DF +AKSALSFT
Sbjct: 708 TDLVFDVQFMLKSAMQYYGFSLEVRTEARKVHNLFFDILKIAFPDTDFQDAKSALSFTGP 887
Query: 1969 ISANAGASS-KQATVFPSKRKRGKNDMETDPTPTQKPLQRGST 2010
ISA SS +Q V KR R N++E D P+Q+ LQRGST
Sbjct: 888 ISAPTMVSSPRQVAVGQGKRHRLVNEVEPDSHPSQRQLQRGST 1016
>TC90445 homologue to PIR|C84507|C84507 hypothetical protein At2g13370
[imported] - Arabidopsis thaliana, partial (29%)
Length = 1672
Score = 107 bits (267), Expect(4) = 3e-53
Identities = 69/176 (39%), Positives = 94/176 (53%), Gaps = 10/176 (5%)
Frame = +1
Query: 1117 PPKVSIVLRCRMSAFQSAIYDWIKSTGTLRLNPEEEQSRMEKSPLYQAKQYKTLNNRCME 1176
PPK+S R MS Q Y WI LN +++ +L N +E
Sbjct: 664 PPKLSASFRVEMSPLQKQYYKWILERNFQNLNKGVRGNQV------------SLLNIVVE 807
Query: 1177 LRKTCNHPLL---------NYPFFSDLSK-DFMVKCCGKLWMLDRILIKLQRTGHRVLLF 1226
L+K CNHP L SD SK + +V GKL +LD++L++L T HRVL+F
Sbjct: 808 LKKCCNHPFLFESADHGYGGDSGGSDNSKLERIVFSSGKLVILDKLLVRLHETKHRVLIF 987
Query: 1227 STMTKLLDILEEYLQWRRLVYRRIDGTTALEDRESAIVDFNSPNSDCFIFLLSIRA 1282
S M ++LDIL +YL R ++R+DG+T E R+ A+ FN+P SD F FLLS RA
Sbjct: 988 SQMVRMLDILAQYLSLRGFQFQRLDGSTKSELRQQAMEHFNAPGSDDFCFLLSTRA 1155
Score = 70.5 bits (171), Expect(4) = 3e-53
Identities = 51/167 (30%), Positives = 79/167 (46%), Gaps = 32/167 (19%)
Frame = +2
Query: 925 ADEMGLGKTVQVMALIAYLMEFKGNYGPHLIIVPNAVLVNWKCAFNP------------- 971
ADEMGLGKTVQ ++++ +L + +GP L++VP + L NW F
Sbjct: 41 ADEMGLGKTVQSVSMLGFLQNAQQIHGPFLVVVPLSTLSNWAKEFRKWLPDMNIIVYVGT 220
Query: 972 ----ENCIDH---------------AE*IAYMVTIGVMHFLCWIEGSSVKIIFSTQRMKD 1012
E C + A Y V + L I+ + + ++ R+K+
Sbjct: 221 RASREVCQQYEFYNDKKPGKPIKFNALLTTYEVILKDKAVLSKIKWNYL-MVDEAHRLKN 397
Query: 1013 RESVLARDLDRYRCHRRLLLTGTPLQNDLKELWSLLNLLLPEVFDNK 1059
E+ L L + +LL+TGTPLQN ++ELW+LL+ L P F +K
Sbjct: 398 SEAQLYTSLLEFSTKNKLLITGTPLQNSVEELWALLHFLDPTKFKSK 538
Score = 54.3 bits (129), Expect(4) = 3e-53
Identities = 24/42 (57%), Positives = 34/42 (80%)
Frame = +2
Query: 1284 GRGLNLQSADTVVIYDPDPNPKNEEQAVARAHRIGQKREVKV 1325
G G+NL +ADTV+I+D D NP+N+ QA++RAHRIGQ+ V +
Sbjct: 1160 GLGINLATADTVIIFDSDWNPQNDLQAMSRAHRIGQQDVVNI 1285
Score = 38.5 bits (88), Expect(4) = 3e-53
Identities = 20/55 (36%), Positives = 30/55 (54%), Gaps = 5/55 (9%)
Frame = +3
Query: 1075 PNQNAENDWLETEKKVIIIHR-----LHQILEPFMLRRRVEEVEGSLPPKVSIVL 1124
PN A +++ K + H LH L P +LRR +++VE SLPPK+ +L
Sbjct: 522 PNSRARMKFVQNYKNLSSFHENELANLHMELRPHILRRVIKDVEKSLPPKIERIL 686
>TC80039 homologue to GP|6437558|gb|AAF08585.1| putative ATPase (ISW2-like)
{Arabidopsis thaliana}, partial (47%)
Length = 1840
Score = 185 bits (469), Expect = 2e-46
Identities = 108/265 (40%), Positives = 151/265 (56%), Gaps = 3/265 (1%)
Frame = +1
Query: 1036 PLQNDLKELWSLLNLLLPEVFDNKKAFNDWFSKPFQKEDPNQNAENDWLETEKKVIIIHR 1095
PLQN+L ELWSLLN LLPE+F + + F++WF + END E ++ +
Sbjct: 1120 PLQNNLHELWSLLNFLLPEIFSSAETFDEWFQI---------SGENDQQE------VVQQ 1254
Query: 1096 LHQILEPFMLRRRVEEVEGSLPPKVSIVLRCRMSAFQSAIYDWIKSTGTLRLNPEEEQSR 1155
LH++L PF+LRR +VE LPPK +L+ MS Q Y + +N E+ R
Sbjct: 1255 LHKVLRPFLLRRLKSDVEKGLPPKKETILKVGMSQMQKQYYKALLQKDLEVVNAGGERKR 1434
Query: 1156 MEKSPLYQAKQYKTLNNRCMELRKTCNHPLLNY---PFFSDLSKDFMVKCCGKLWMLDRI 1212
L N M+LRK CNHP L P + D ++ GK+ ++D++
Sbjct: 1435 --------------LLNIAMQLRKCCNHPYLFQGAEPGPPYTTGDHIITSAGKMVLMDKL 1572
Query: 1213 LIKLQRTGHRVLLFSTMTKLLDILEEYLQWRRLVYRRIDGTTALEDRESAIVDFNSPNSD 1272
L KL+ RVL+FS MT+LLDILE+YL +R Y RIDG T +DR+++I FN P S+
Sbjct: 1573 LPKLKERDSRVLIFSQMTRLLDILEDYLMFRGYQYCRIDGNTGGDDRDASIEAFNKPGSE 1752
Query: 1273 CFIFLLSIRAAGRGLNLQSADTVVI 1297
F+FLLS RA G G+NL +AD V++
Sbjct: 1753 KFVFLLSTRAGGLGINLATADVVIL 1827
Score = 98.2 bits (243), Expect = 3e-20
Identities = 42/80 (52%), Positives = 61/80 (75%)
Frame = +1
Query: 886 NEKVLRQPSMLRAGTLREYQLVGLQWMLSLYNNKLNGILADEMGLGKTVQVMALIAYLME 945
N +++ QPS ++ G +R+YQL GL W++ LY N +NGILADEMGLGKT+Q ++L+ YL E
Sbjct: 598 NTRLVTQPSCIQ-GKMRDYQLAGLNWLIRLYENGINGILADEMGLGKTLQTISLLGYLHE 774
Query: 946 FKGNYGPHLIIVPNAVLVNW 965
F+G GPH+++ P + L NW
Sbjct: 775 FRGITGPHMVVAPKSTLGNW 834
>CB892431 similar to GP|10039641|gb| Orf122 {Chlorobium tepidum}, partial (60%)
Length = 645
Score = 158 bits (399), Expect = 2e-38
Identities = 80/105 (76%), Positives = 89/105 (84%)
Frame = +1
Query: 1788 RKVANASKLHVSPKSSRLNCTSAPSEDNDEHSRERLKGKPNNLRGSSAHVTNMTEIIQRR 1847
++VAN SKLH SPK +RL SAPSED EHSRE +GKP N GSSAH + MTEIIQRR
Sbjct: 340 QRVANTSKLHSSPKPTRL---SAPSEDGGEHSRESWEGKPINSSGSSAHGSRMTEIIQRR 510
Query: 1848 CKSVISKLQRRIDKEGHQIVPLLTDLWKRIENSGFAGGSGNNLLD 1892
CK+VISKLQRRIDKEGHQIVPLLTDLWK IENSG++GGSGNNL+D
Sbjct: 511 CKNVISKLQRRIDKEGHQIVPLLTDLWKEIENSGYSGGSGNNLVD 645
>TC87478 homologue to PIR|G84897|G84897 hypothetical protein At2g46020
[imported] - Arabidopsis thaliana, partial (1%)
Length = 980
Score = 142 bits (358), Expect = 1e-33
Identities = 84/154 (54%), Positives = 102/154 (65%), Gaps = 14/154 (9%)
Frame = +2
Query: 2000 PTQKPLQRGSTSNSESGRIKVQLPQKASRTGSGSGSA-REQLQQ--DSPSLLTHPGDLVV 2056
P+Q+ LQRGS S+ E+ RI+V++P K SR+G GSGS+ REQ QQ DSP LLTHPG+LVV
Sbjct: 41 PSQRQLQRGSASSGENSRIRVRVPPKESRSGYGSGSSIREQPQQQDDSPPLLTHPGELVV 220
Query: 2057 CKKKRNERGDKSSVKHRIGSAGPVSPPKIVVHTVLAERSPTPGSGSTPR----------- 2105
CKK+RNER +KS VK R GPVSP +PG+GS P+
Sbjct: 221 CKKRRNER-EKSLVKSR---TGPVSPSM-----------RSPGAGSVPKDVRLTQQTQGW 355
Query: 2106 AGHAHTSNGSGGSVGWANPVKRMRTDSGKRRPSH 2139
G + +GGSVGWANPVKR+RTDSGKRRPSH
Sbjct: 356 TGQPSSQQPNGGSVGWANPVKRLRTDSGKRRPSH 457
>BG645739 similar to PIR|T45808|T4 helicase-like protein - Arabidopsis
thaliana, partial (17%)
Length = 818
Score = 136 bits (343), Expect = 8e-32
Identities = 83/250 (33%), Positives = 132/250 (52%), Gaps = 31/250 (12%)
Frame = +2
Query: 923 ILADEMGLGKTVQVMALIAYLMEFKGNYGPHLIIVPNAVLVNW----------------- 965
ILADEMGLGKT+Q M +A+L E K +GP LI+ P +VL NW
Sbjct: 2 ILADEMGLGKTIQAMVFLAHLAEEKNIWGPFLIVAPASVLNNWNEELERFCPELKVLPYW 181
Query: 966 ---------KCAFNPENCID-----HAE*IAYMVTIGVMHFLCWIEGSSVKIIFSTQRMK 1011
+ + NP++ H +Y + + + ++ + ++ Q +K
Sbjct: 182 GGLSERTVLRKSMNPKDLYRREAKFHILITSYQLLVSDEKYFRRVKWQYM-VLDEAQAIK 358
Query: 1012 DRESVLARDLDRYRCHRRLLLTGTPLQNDLKELWSLLNLLLPEVFDNKKAFNDWFSKPFQ 1071
S+ + L + C RLLLTGTP+QN++ ELW+LL+ ++P +FD+ + FN+WFSK +
Sbjct: 359 SSNSIRWKTLLSFNCRNRLLLTGTPVQNNMAELWALLHFIMPTLFDSHEQFNEWFSKGIE 538
Query: 1072 KEDPNQNAENDWLETEKKVIIIHRLHQILEPFMLRRRVEEVEGSLPPKVSIVLRCRMSAF 1131
+AE+ E + ++RLH I++PFMLRR ++V L K I + C++S+
Sbjct: 539 -----NHAEHGGTLNEHQ---LNRLHSIIKPFMLRRVKKDVVSELTSKTEITVHCKLSSR 694
Query: 1132 QSAIYDWIKS 1141
Q A Y IK+
Sbjct: 695 QQAFYSSIKN 724
>CA990634 homologue to PIR|G84897|G84 hypothetical protein At2g46020 [imported]
- Arabidopsis thaliana, partial (1%)
Length = 549
Score = 135 bits (339), Expect = 2e-31
Identities = 63/63 (100%), Positives = 63/63 (100%)
Frame = +3
Query: 2077 AGPVSPPKIVVHTVLAERSPTPGSGSTPRAGHAHTSNGSGGSVGWANPVKRMRTDSGKRR 2136
AGPVSPPKIVVHTVLAERSPTPGSGSTPRAGHAHTSNGSGGSVGWANPVKRMRTDSGKRR
Sbjct: 3 AGPVSPPKIVVHTVLAERSPTPGSGSTPRAGHAHTSNGSGGSVGWANPVKRMRTDSGKRR 182
Query: 2137 PSH 2139
PSH
Sbjct: 183 PSH 191
>TC84487 similar to PIR|T52301|T52301 GYMNOS/PICKLE protein [imported] -
Arabidopsis thaliana, partial (14%)
Length = 612
Score = 132 bits (332), Expect = 1e-30
Identities = 73/162 (45%), Positives = 103/162 (63%), Gaps = 5/162 (3%)
Frame = +3
Query: 1169 TLNNRCMELRKTCNHPLLNY---PFFSDLSKDF--MVKCCGKLWMLDRILIKLQRTGHRV 1223
+L N MELRK C H + P D + F +++ GKL +LD++++KL+ GHRV
Sbjct: 96 SLINVVMELRKLCCHAYMLEGVEPDIDDPKEAFKQLLESSGKLHLLDKMMVKLKEQGHRV 275
Query: 1224 LLFSTMTKLLDILEEYLQWRRLVYRRIDGTTALEDRESAIVDFNSPNSDCFIFLLSIRAA 1283
L++S +LD+LE+Y +++ Y RIDG +R+ I FN+ NS F FLLS RA
Sbjct: 276 LIYSQFQHMLDLLEDYCSYKKWHYERIDGKVGGAERQIRIDRFNAKNSSRFCFLLSTRAG 455
Query: 1284 GRGLNLQSADTVVIYDPDPNPKNEEQAVARAHRIGQKREVKV 1325
G G+NL +ADTVVIYD D NP + QA+ARAHR+GQ +V +
Sbjct: 456 GLGINLATADTVVIYDRDWNPHADLQAMARAHRLGQTNKVLI 581
>BG452179 homologue to GP|15528650|d putative DNA-dependent ATPase {Oryza
sativa (japonica cultivar-group)}, partial (19%)
Length = 667
Score = 115 bits (288), Expect(2) = 2e-30
Identities = 76/214 (35%), Positives = 108/214 (49%), Gaps = 24/214 (11%)
Frame = +3
Query: 943 LMEFKGNYGPHLIIVPNAVLVNW--------------KCAFNPENCIDHAE*--IAYMVT 986
L EF+G GPH+++ P + L NW K NPE E +A
Sbjct: 69 LHEFRGIKGPHMVVAPKSTLGNWMNEIRRFCPILRAVKFLGNPEERRHIREDLLVAGKFD 248
Query: 987 IGVMHFLCWIEGSSVK--------IIFSTQRMKDRESVLARDLDRYRCHRRLLLTGTPLQ 1038
+ V F I+ S II R+K+ S+L++ + Y + RLL+TGTPLQ
Sbjct: 249 VCVTSFEMAIKEKSTLRRFSWRYIIIDEAHRIKNENSLLSKTMRIYNTNYRLLITGTPLQ 428
Query: 1039 NDLKELWSLLNLLLPEVFDNKKAFNDWFSKPFQKEDPNQNAENDWLETEKKVIIIHRLHQ 1098
N+L ELWSLLN LLPE+F + + F++WF + END E ++ +LH+
Sbjct: 429 NNLHELWSLLNFLLPEIFSSAETFDEWF---------QISGENDQQE------VVQQLHK 563
Query: 1099 ILEPFMLRRRVEEVEGSLPPKVSIVLRCRMSAFQ 1132
+L PF+LRR +VE LPPK +L+ MS Q
Sbjct: 564 VLRPFLLRRLKSDVEKGLPPKKETILKVGMSQLQ 665
Score = 37.7 bits (86), Expect(2) = 2e-30
Identities = 16/22 (72%), Positives = 20/22 (90%)
Frame = +2
Query: 921 NGILADEMGLGKTVQVMALIAY 942
NGILADEMGLGKT+Q ++L+ Y
Sbjct: 2 NGILADEMGLGKTLQTISLMGY 67
>TC84105 homologue to PIR|C84507|C84507 hypothetical protein At2g13370
[imported] - Arabidopsis thaliana, partial (15%)
Length = 834
Score = 125 bits (313), Expect = 2e-28
Identities = 76/215 (35%), Positives = 124/215 (57%), Gaps = 8/215 (3%)
Frame = +1
Query: 1210 DRILIKLQRTGHRVLLFSTMTKLLDILEEYLQWRRLVYRRIDGTTALEDRESAIVDFNSP 1269
D++L++L T R+L+FS M ++LDIL +Y+ R ++R+DG+T E R+ A+ FN+P
Sbjct: 1 DKLLVRLHETKFRILIFSQMVRMLDILAQYMSLRGFQFQRLDGSTKSELRQQAMDHFNAP 180
Query: 1270 NSDCFIFLLSIRAAGRGLNLQSADTVVIYDPDPNPKNEEQAVARAHRIGQKREVKVIYM- 1328
SD F FLLS RA G G+NL +ADTV+I+D D NP+N+ QA++RAHRIGQ REV IY
Sbjct: 181 GSDDFCFLLSTRAGGLGINLATADTVIIFDSDWNPQNDLQAMSRAHRIGQ-REVVNIYRF 357
Query: 1329 -------EAVVDKISSHQKEDEMRIGGTIDMEDELAGKDRYIGSIESLIRSNIQQYKIDM 1381
E ++++ D + I ++ E +L K+ G ++ +
Sbjct: 358 VTSKSVEEDILERAKKKMVLDHLVI-QKLNAEGKLEKKEAKKGG-SFFDKNELSAILRFG 531
Query: 1382 ADEVINAGRFDQRTTHEERRLTLETLLHDEERCQE 1416
A+E++ R D+ + + ++ +L E+ +E
Sbjct: 532 AEELVKEERNDEESKKRLLSMDIDEILERAEKVEE 636
>BQ122721 similar to GP|8843733|dbj SWI2/SNF2-like protein {Arabidopsis
thaliana}, partial (19%)
Length = 650
Score = 107 bits (267), Expect = 5e-23
Identities = 62/158 (39%), Positives = 87/158 (54%), Gaps = 21/158 (13%)
Frame = +1
Query: 1221 HRVLLFSTMTKLLDILEEYLQWRRLVYRRIDGTTALEDRESAIVDFNSPNSDCFIFLLSI 1280
H+VL+FS TK+LDI++ Y + RIDG+ L+DR+ I DFN S+C IFLLS
Sbjct: 16 HKVLIFSQWTKVLDIMDYYFSEKGFEVCRIDGSVKLDDRKRQIQDFNDTTSNCRIFLLST 195
Query: 1281 RAAGRGLNLQSADTVVIYDPDPNPKNEEQAVARAHRIGQKREVKVIYM------------ 1328
RA G G+NL +ADT ++YD D NP+ + QA+ R HRIGQ + V V +
Sbjct: 196 RAGGLGINLTAADTCILYDSDWNPQMDLQAMDRCHRIGQTKPVHVYRLATAQSVEGRMLK 375
Query: 1329 ---------EAVVDKISSHQKEDEMRIGGTIDMEDELA 1357
V++K HQ+ + I ++ ED LA
Sbjct: 376 RAFSKLKLEHVVIEKGQFHQERTKPSIMDEMEEEDVLA 489
>BE205152 similar to GP|19347965|gb putative helicase {Arabidopsis thaliana},
partial (27%)
Length = 626
Score = 107 bits (266), Expect = 6e-23
Identities = 54/108 (50%), Positives = 76/108 (70%)
Frame = +1
Query: 1204 GKLWMLDRILIKLQRTGHRVLLFSTMTKLLDILEEYLQWRRLVYRRIDGTTALEDRESAI 1263
GKL LD +L +L+ HRVLLF+ MTK+L+ILE+Y+ +R+ Y R+DG+T++ DR +
Sbjct: 301 GKLQTLDILLKRLRAGNHRVLLFAQMTKMLNILEDYMNYRKYKYCRLDGSTSIHDRRDMV 480
Query: 1264 VDFNSPNSDCFIFLLSIRAAGRGLNLQSADTVVIYDPDPNPKNEEQAV 1311
DF SD F+FLLS RA G G+NL +ADTV+ Y+ D NP + QA+
Sbjct: 481 RDFQH-RSDIFVFLLSTRAGGLGINLTAADTVIFYESDWNPTLDLQAM 621
>TC83002 similar to GP|14532630|gb|AAK64043.1 putative DNA-binding protein
{Arabidopsis thaliana}, partial (18%)
Length = 1088
Score = 84.0 bits (206), Expect = 6e-16
Identities = 48/140 (34%), Positives = 80/140 (56%), Gaps = 3/140 (2%)
Frame = +3
Query: 1222 RVLLFSTMTKLLDILEEYLQWRRLVYRRIDGTTALEDRESAIVDFNSPNSDCFIFLLSIR 1281
+ ++FS T +LD++E ++ + YRR+DG L R+ A+ DFN+ + + + L+S++
Sbjct: 453 KAIIFSQWTSMLDLVETSMEQSGVKYRRLDGRMTLTARDRAVKDFNT-DPEITVMLMSLK 629
Query: 1282 AAGRGLNLQSADTVVIYDPDPNPKNEEQAVARAHRIGQKREV---KVIYMEAVVDKISSH 1338
A GLN+ +A V++ D NP E+QA+ RAHRIGQ R V ++ + V D+I +
Sbjct: 630 AGNLGLNMVAACHVILLDLWWNPTTEDQAIDRAHRIGQTRPVTVTRITIKDTVEDRILAL 809
Query: 1339 QKEDEMRIGGTIDMEDELAG 1358
Q+E + ED G
Sbjct: 810 QEEKRKMVASAFG-EDHAGG 866
>BG584027 similar to GP|10178052|db helicase-like transcription factor-like
protein {Arabidopsis thaliana}, partial (15%)
Length = 830
Score = 73.9 bits (180), Expect = 6e-13
Identities = 48/134 (35%), Positives = 73/134 (53%), Gaps = 4/134 (2%)
Frame = +3
Query: 1217 QRTGHRVLLFSTMTKLLDILEEYLQWRRLVYRRIDGTTALEDRESAIVDFN-SPNSDCFI 1275
Q + ++FS K+L +LEE L+ R+DGT + R I F S + I
Sbjct: 246 QNPATKSVVFSQFRKMLLLLEEPLKAAGFKTLRLDGTMNAKQRAQVIEQFQLSEVDEPMI 425
Query: 1276 FLLSIRAAGRGLNLQSADTVVIYDPDPNPKNEEQAVARAHRIGQKREVKVIYM---EAVV 1332
L S+RA+ G+NL +A V + +P NP EEQA+ R HRIGQK EVK++ + ++
Sbjct: 426 LLASLRASSTGINLTAASRVYLMEPWWNPAVEEQAMDRVHRIGQKEEVKIVRLIAKNSIE 605
Query: 1333 DKISSHQKEDEMRI 1346
+KI Q++ + I
Sbjct: 606 EKILMLQEKKKKTI 647
>TC90905 similar to PIR|H84432|H84432 probable helicase [imported] -
Arabidopsis thaliana, partial (20%)
Length = 904
Score = 70.5 bits (171), Expect = 7e-12
Identities = 34/67 (50%), Positives = 46/67 (67%), Gaps = 1/67 (1%)
Frame = +2
Query: 901 LREYQLVGLQWMLSLYNNKLNG-ILADEMGLGKTVQVMALIAYLMEFKGNYGPHLIIVPN 959
L+ YQLVG+ ++L LY ++ G ILADEMGLGKTVQ + + L + GPHLI+ P
Sbjct: 701 LKPYQLVGVNFLLLLYRKRIGGAILADEMGLGKTVQAITYLTLLNHLHNDSGPHLIVCPA 880
Query: 960 AVLVNWK 966
+VL NW+
Sbjct: 881 SVLENWE 901
>TC84267 weakly similar to PIR|S41478|S41478 DNA repair protein rad8 - fission
yeast (Schizosaccharomyces pombe), partial (5%)
Length = 751
Score = 67.8 bits (164), Expect = 4e-11
Identities = 43/125 (34%), Positives = 69/125 (54%), Gaps = 3/125 (2%)
Frame = +2
Query: 1218 RTGHRVLLFSTMTKLLDILEEYLQWRRLVYRRIDGTTALEDRESAIVDFNSPNSDCFIFL 1277
R R ++FS T+ L ++E YL+ + + RIDG +L +R++ + F + D + +
Sbjct: 122 RLSQRSIIFSCWTRTLRLVERYLRAAGIEHLRIDGDCSLPERQARLKQF-ADGDDKRVLI 298
Query: 1278 LSIRAAGRGLNLQSADTVVIYDPDPNPKNEEQAVARAHRIGQKREVKV---IYMEAVVDK 1334
++ GLNL A+ V I + NP E QA+ARA R+GQ REV V + + V +
Sbjct: 299 MTTGTGAFGLNLTCANRVFIVELQWNPSVENQAIARAIRLGQGREVLVTRYVMRDTVEVE 478
Query: 1335 ISSHQ 1339
+SS Q
Sbjct: 479 MSSQQ 493
>BF632064 similar to PIR|H84432|H84 probable helicase [imported] - Arabidopsis
thaliana, partial (26%)
Length = 646
Score = 66.6 bits (161), Expect = 1e-10
Identities = 50/172 (29%), Positives = 84/172 (48%), Gaps = 5/172 (2%)
Frame = +3
Query: 1027 HRRLLLTGTPLQNDLKELWSLLNLLLPEVFDNKKAFNDWFSKPFQKEDPNQNAENDWLET 1086
++RL+LTGTPLQNDL ELWS+L ++P++F ++ K ED +
Sbjct: 3 NQRLMLTGTPLQNDLHELWSMLEFMMPDIFASEDVD---LKKLLGAEDKD---------- 143
Query: 1087 EKKVIIIHRLHQILEPFMLRRRVEEVEGSLPPKVS-----IVLRCRMSAFQSAIYDWIKS 1141
+ R+ IL PF+LRR +V L K I+ + + A++ AI ++ ++
Sbjct: 144 -----LTSRMKSILGPFILRRLKSDVMQQLVRKTQKVQYVIMEKQQEHAYKEAIEEY-RA 305
Query: 1142 TGTLRLNPEEEQSRMEKSPLYQAKQYKTLNNRCMELRKTCNHPLLNYPFFSD 1193
RL + S + + + + +NN ++ RK NHPLL ++D
Sbjct: 306 VSQARLT---KCSDLNPKNVLEVLPRRQINNYFVQFRKIANHPLLIRRIYND 452
>TC91548 similar to PIR|D86185|D86185 hypothetical protein [imported] -
Arabidopsis thaliana, complete
Length = 611
Score = 64.3 bits (155), Expect = 5e-10
Identities = 39/111 (35%), Positives = 59/111 (53%), Gaps = 3/111 (2%)
Frame = +2
Query: 1252 GTTALEDRESAIVDFNSPNSDCFIFLLSIRAAGRGLNLQSADTVVIYDPDPNPKNEEQAV 1311
G+ L R++AI F + DC IFL+S++A G LNL A V + DP NP E QA
Sbjct: 14 GSMTLTARDNAIKKFTD-DPDCKIFLMSLKAGGVALNLTVASHVFLMDPWWNPAVERQAQ 190
Query: 1312 ARAHRIGQKREVKVIYM---EAVVDKISSHQKEDEMRIGGTIDMEDELAGK 1359
R HRIGQ + ++++ + ++I Q++ E+ G + E GK
Sbjct: 191 DRIHRIGQYKPIRIVRFVIENTIEERILKLQEKKELVFEGAVGGSSEALGK 343
>BI311783 homologue to PIR|T47587|T4 TATA box binding protein (TBP) associated
factor (TAF)-like protein - Arabidopsis thaliana, partial
(6%)
Length = 788
Score = 54.7 bits (130), Expect = 4e-07
Identities = 39/128 (30%), Positives = 63/128 (48%)
Frame = +2
Query: 1277 LLSIRAAGRGLNLQSADTVVIYDPDPNPKNEEQAVARAHRIGQKREVKVIYMEAVVDKIS 1336
LL+ G GLNL SADT+V + D NP + QA+ RAHR+GQK KV+ + ++ +
Sbjct: 5 LLTTHVGGLGLNLTSADTLVFVEHDWNPMRDLQAMDRAHRLGQK---KVVNVHRLIMR-- 169
Query: 1337 SHQKEDEMRIGGTIDMEDELAGKDRYIGSIESLIRSNIQQYKIDMADEVINAGRFDQRTT 1396
G++E + S +Q++K+ +A+ VINA +T
Sbjct: 170 ---------------------------GTLEEKVMS-LQRFKVSVANAVINAENASLKTM 265
Query: 1397 HEERRLTL 1404
+ ++ L L
Sbjct: 266 NTDQLLDL 289
>TC85956 homologue to GP|8885593|dbj|BAA97523.1 1 4-benzoquinone
reductase-like; Trp repressor binding protein-like
{Arabidopsis thaliana}, partial (98%)
Length = 1976
Score = 54.3 bits (129), Expect = 5e-07
Identities = 47/174 (27%), Positives = 85/174 (48%), Gaps = 9/174 (5%)
Frame = +1
Query: 1181 CNHPLLNYPFFSDLSK-DFMVKCCGKLWMLDRILIKLQRTGHRVLLF----STMTKLLDI 1235
C+ L SDL K F +K K+ + ++ ++ + +VL+F + + +
Sbjct: 25 CSQKFLTKEQLSDLDKYKFDLKIGSKVRFVLSLIYRVVKN-EKVLIFCHNIAPVRLFQEY 201
Query: 1236 LEEYLQWRR----LVYRRIDGTTALEDRESAIVDFNSPNSDCFIFLLSIRAAGRGLNLQS 1291
E+Y W++ LV + G L +R + F P I L SI A G++L +
Sbjct: 202 FEKYFGWQKGREVLV---LTGELELFERGKIMDKFEEPGGVSKILLASITACAEGISLTA 372
Query: 1292 ADTVVIYDPDPNPKNEEQAVARAHRIGQKREVKVIYMEAVVDKISSHQKEDEMR 1345
A V++ D + NP +QA+ARA R GQ+ K++Y+ ++ ++ +ED+ R
Sbjct: 373 ASRVIMLDSEWNPSKTKQAIARAFRPGQQ---KMVYVYQLL--VTGSLEEDKYR 519
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.313 0.130 0.363
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 55,247,292
Number of Sequences: 36976
Number of extensions: 732138
Number of successful extensions: 3432
Number of sequences better than 10.0: 77
Number of HSP's better than 10.0 without gapping: 3283
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3397
length of query: 2139
length of database: 9,014,727
effective HSP length: 111
effective length of query: 2028
effective length of database: 4,910,391
effective search space: 9958272948
effective search space used: 9958272948
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 66 (30.0 bits)
Medicago: description of AC139601.1


