

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
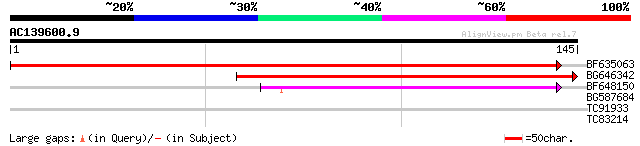
Query= AC139600.9 - phase: 0 /pseudo
(145 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BF635063 weakly similar to PIR|F84486|F84 probable retroelement ... 231 1e-61
BG646342 weakly similar to PIR|F84486|F84 probable retroelement ... 134 1e-32
BF648150 similar to GP|14586969|gb| pol polyprotein {Citrus x pa... 45 1e-05
BG587684 similar to GP|12858190|dbj evidence:NAS~putative~unclas... 30 0.44
TC91933 similar to GP|7270830|emb|CAB80511.1 protein kinase like... 26 4.9
TC83214 similar to SP|O49204|KAPS_CATRO Adenylylsulfate kinase ... 26 6.4
>BF635063 weakly similar to PIR|F84486|F84 probable retroelement pol
polyprotein [imported] - Arabidopsis thaliana, partial
(4%)
Length = 677
Score = 231 bits (588), Expect = 1e-61
Identities = 120/141 (85%), Positives = 129/141 (91%)
Frame = -2
Query: 1 TMGSKWDIEKFTGDNDFGLLKVKMEAMLIQQKCEKALKGKGSLPVTMSRAEKTEMVDKAR 60
TMGSK DIEKFTGDNDFGL KVKMEA+LIQQKCEKALKG+ SLPVTMSRAEKTEMVDKAR
Sbjct: 571 TMGSKRDIEKFTGDNDFGLWKVKMEAVLIQQKCEKALKGEVSLPVTMSRAEKTEMVDKAR 392
Query: 61 SVIVLCLGDKVLRKVAKESTAASMWPKLEALYMTKSLTHQQFLKQ*LYSFKMVKSKAIME 120
S IVLCLGDKVLR+VAKE TAASMW KL +LYMTKSL H+QFLKQ LYSF+MV+SKAIME
Sbjct: 391 SAIVLCLGDKVLREVAKERTAASMWAKL*SLYMTKSLAHRQFLKQQLYSFRMVESKAIME 212
Query: 121 LLVEFNKIIGDLENIEVRLED 141
L EFNKI+ DLENIEV+LED
Sbjct: 211 QLTEFNKILDDLENIEVQLED 149
>BG646342 weakly similar to PIR|F84486|F84 probable retroelement pol
polyprotein [imported] - Arabidopsis thaliana, partial
(4%)
Length = 599
Score = 134 bits (338), Expect = 1e-32
Identities = 72/87 (82%), Positives = 75/87 (85%)
Frame = +2
Query: 59 ARSVIVLCLGDKVLRKVAKESTAASMWPKLEALYMTKSLTHQQFLKQ*LYSFKMVKSKAI 118
ARS IVLCLGDKVLR+VAKE TA SM KLE LYMTKSL H+QFLKQ LYSFKMV+SKAI
Sbjct: 2 ARSAIVLCLGDKVLREVAKEPTATSMCAKLEYLYMTKSLAHRQFLKQQLYSFKMVESKAI 181
Query: 119 MELLVEFNKIIGDLENIEVRLEDVDAL 145
ELLVEFNKIIGDLENIEV LED AL
Sbjct: 182 TELLVEFNKIIGDLENIEVHLEDAGAL 262
>BF648150 similar to GP|14586969|gb| pol polyprotein {Citrus x paradisi},
partial (3%)
Length = 658
Score = 45.1 bits (105), Expect = 1e-05
Identities = 23/84 (27%), Positives = 43/84 (50%), Gaps = 7/84 (8%)
Frame = +2
Query: 65 LCLG-------DKVLRKVAKESTAASMWPKLEALYMTKSLTHQQFLKQ*LYSFKMVKSKA 117
+CLG D + +A +W KLE YM + T ++FL ++KMV +K+
Sbjct: 197 ICLGHILNGMSDSLFDIYQSSPSAKDLWDKLETRYMREDATSKKFLVSHFNNYKMVDNKS 376
Query: 118 IMELLVEFNKIIGDLENIEVRLED 141
+ME L E +I+ + + + +++
Sbjct: 377 VMEQLYEIERILNNYKQHNMNMDE 448
>BG587684 similar to GP|12858190|dbj evidence:NAS~putative~unclassifiable
{Mus musculus}, partial (14%)
Length = 819
Score = 29.6 bits (65), Expect = 0.44
Identities = 11/20 (55%), Positives = 16/20 (80%)
Frame = +3
Query: 2 MGSKWDIEKFTGDNDFGLLK 21
M +KWDIE F G+NDF +++
Sbjct: 735 MDTKWDIEIFIGENDFRVVE 794
>TC91933 similar to GP|7270830|emb|CAB80511.1 protein kinase like protein
{Arabidopsis thaliana}, partial (9%)
Length = 832
Score = 26.2 bits (56), Expect = 4.9
Identities = 14/35 (40%), Positives = 21/35 (60%)
Frame = -1
Query: 21 KVKMEAMLIQQKCEKALKGKGSLPVTMSRAEKTEM 55
K K E L+ K +K KGK + + SRAE+T++
Sbjct: 424 KTKEERRLL*LKKKKKKKGKSRV*LDFSRAERTKL 320
>TC83214 similar to SP|O49204|KAPS_CATRO Adenylylsulfate kinase chloroplast
precursor (EC 2.7.1.25) (APS kinase), partial (23%)
Length = 603
Score = 25.8 bits (55), Expect = 6.4
Identities = 16/63 (25%), Positives = 30/63 (47%), Gaps = 1/63 (1%)
Frame = -3
Query: 43 LPVTMSRAEKTEMVDKARSVIVLCLGDKVLRKVAKESTAASMWPKLE-ALYMTKSLTHQQ 101
+P+ + +D +RS + +G +L + + STA ++W KL + T + H
Sbjct: 292 IPLYFGILLSSSAIDDSRSSCAM-IGFNILLRQRRSSTALNLWRKLTIPNFSTVEVPHSM 116
Query: 102 FLK 104
F K
Sbjct: 115 FWK 107
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.322 0.135 0.377
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,142,500
Number of Sequences: 36976
Number of extensions: 28796
Number of successful extensions: 120
Number of sequences better than 10.0: 12
Number of HSP's better than 10.0 without gapping: 120
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 120
length of query: 145
length of database: 9,014,727
effective HSP length: 87
effective length of query: 58
effective length of database: 5,797,815
effective search space: 336273270
effective search space used: 336273270
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 53 (25.0 bits)
Medicago: description of AC139600.9


