

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC139600.7 - phase: 0
(320 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
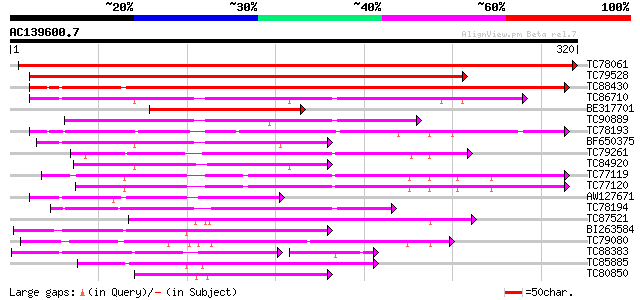
Score E
Sequences producing significant alignments: (bits) Value
TC78061 weakly similar to PIR|T10824|T10824 auxin-induced protei... 435 e-122
TC79528 similar to GP|15808674|gb|AAL06644.1 putative quinone ox... 352 1e-97
TC88430 weakly similar to PIR|T10824|T10824 auxin-induced protei... 285 2e-77
TC86710 similar to GP|15724256|gb|AAL06521.1 AT4g13010/F25G13_10... 137 7e-33
BE317701 similar to GP|14532287|gb| quinone oxidoreductase-like ... 122 2e-28
TC90889 similar to GP|15724256|gb|AAL06521.1 AT4g13010/F25G13_10... 110 6e-25
TC78193 similar to PIR|T05166|T05166 quinone reductase homolog F... 108 2e-24
BF650375 similar to GP|15724256|gb AT4g13010/F25G13_100 {Arabido... 105 2e-23
TC79261 similar to PIR|T49047|T49047 quinone reductase-like prot... 100 6e-22
TC84920 similar to GP|15724256|gb|AAL06521.1 AT4g13010/F25G13_10... 96 2e-20
TC77119 similar to PIR|D96533|D96533 ARP protein [imported] - Ar... 96 2e-20
TC77120 similar to PIR|D96533|D96533 ARP protein [imported] - Ar... 94 7e-20
AW127671 weakly similar to GP|15724256|gb AT4g13010/F25G13_100 {... 88 4e-18
TC78194 similar to PIR|T05166|T05166 quinone reductase homolog F... 86 2e-17
TC87521 similar to SP|Q9ZRF1|MTD_FRAAN Probable mannitol dehydro... 74 7e-14
BI263584 similar to SP|Q9P6C8|ADH1 Alcohol dehydrogenase I (EC 1... 69 2e-12
TC79080 similar to GP|10177043|dbj|BAB10455. alcohol dehydrogena... 68 5e-12
TC88383 similar to GP|21592515|gb|AAM64465.1 nuclear receptor bi... 62 3e-11
TC85885 similar to SP|Q9ZRF1|MTD_FRAAN Probable mannitol dehydro... 60 1e-09
TC80850 homologue to SP|O82515|MTD_MEDSA Probable mannitol dehyd... 57 1e-08
>TC78061 weakly similar to PIR|T10824|T10824 auxin-induced protein (clone
MII-3) - mung bean, complete
Length = 1315
Score = 435 bits (1118), Expect = e-122
Identities = 212/315 (67%), Positives = 261/315 (82%)
Frame = +3
Query: 6 SIPSHTKAWVYSQYGNIEEILKFDSNVPTPHPKEDQVLIKVVAAALNPVDIKRALGHFKD 65
+IP+HTKAW YS++G +ILK N P K+DQVLIKVVAA+LNPVD KR FK+
Sbjct: 99 TIPTHTKAWFYSEHGKASDILKLHPNWSIPQLKDDQVLIKVVAASLNPVDYKRMHALFKE 278
Query: 66 IDSPLPTVPGYDVAGVVVSVGEQVKKFKVGDEVYGDINEITLHNPKTIGTLSEYTVAEEK 125
D LP VPG+DVAG+V+ VG +V KFKVGDE+YGDINE L N K +GTLSEYT+AEE+
Sbjct: 279 TDPHLPIVPGFDVAGIVIKVGSEVVKFKVGDEIYGDINEEGLSNLKILGTLSEYTIAEER 458
Query: 126 VLAHKPSNLSFVEAASLPLAIITAYQGLERVEFSSGKSLLVLGGAGGVGSLVIQLAKHVF 185
+LAHKP NLSF+EAAS+PLA+ TAY+GLER + S+GKS+LVLGGAGGVGS IQLAKHV+
Sbjct: 459 LLAHKPKNLSFIEAASIPLAMETAYEGLERAQLSAGKSILVLGGAGGVGSFAIQLAKHVY 638
Query: 186 EASRIAATASTTKLDFLRKLGADLAIDYTKENYEELTEKFDVVYDAVGDSERAVKAANEG 245
AS+IAAT+ST K++FLRKLG DL IDYTKEN+E+L EKFDVVYD VG+ +RA+KA EG
Sbjct: 639 GASKIAATSSTGKIEFLRKLGVDLPIDYTKENFEDLPEKFDVVYDGVGEVDRAMKAIKEG 818
Query: 246 GKVVTILPPGTPPAIPFLLTSDGAVLEKLQPYLENGKVKPILDPKSPFPFYQTLEAFSYL 305
GKVVTI+PPG PPAI F+LTS G++LEKL+PY E+G++KPILD K+P PF + +EA+SYL
Sbjct: 819 GKVVTIVPPGFPPAIFFVLTSKGSILEKLRPYFESGQLKPILDSKTPVPFSEVIEAYSYL 998
Query: 306 NTNRAIGKIVIHPIP 320
T+RA GK+VI+PIP
Sbjct: 999 ETSRATGKVVIYPIP 1043
>TC79528 similar to GP|15808674|gb|AAL06644.1 putative quinone
oxidoreductase {Fragaria x ananassa}, partial (72%)
Length = 834
Score = 352 bits (903), Expect = 1e-97
Identities = 175/247 (70%), Positives = 203/247 (81%)
Frame = +2
Query: 12 KAWVYSQYGNIEEILKFDSNVPTPHPKEDQVLIKVVAAALNPVDIKRALGHFKDIDSPLP 71
KAW YS+YG+ ++LKFD NVP P K+DQVLIKV AA+LNP+D KR G FK DSPLP
Sbjct: 92 KAWTYSEYGHSVDVLKFDPNVPIPQVKDDQVLIKVAAASLNPIDYKRMEGAFKASDSPLP 271
Query: 72 TVPGYDVAGVVVSVGEQVKKFKVGDEVYGDINEITLHNPKTIGTLSEYTVAEEKVLAHKP 131
T PGYDV+GVVV VG +VKKFKVGDEVYGDIN L PK IG+L+EYT AEE +LAHKP
Sbjct: 272 TAPGYDVSGVVVKVGSEVKKFKVGDEVYGDINVKALEYPKVIGSLAEYTAAEEILLAHKP 451
Query: 132 SNLSFVEAASLPLAIITAYQGLERVEFSSGKSLLVLGGAGGVGSLVIQLAKHVFEASRIA 191
NLSF EAASLPL I TAY+GLER FS+GKS+LVLGGAGGVG+ VIQLAKHV+ AS++A
Sbjct: 452 KNLSFAEAASLPLTIETAYEGLERTGFSAGKSILVLGGAGGVGTHVIQLAKHVYGASKVA 631
Query: 192 ATASTTKLDFLRKLGADLAIDYTKENYEELTEKFDVVYDAVGDSERAVKAANEGGKVVTI 251
AT+ST KL+ L LGADL IDYTKEN+E+L EKFDVV+D VG++E+A KA EGGKVVTI
Sbjct: 632 ATSSTKKLELLSNLGADLPIDYTKENFEDLPEKFDVVFDTVGETEKAFKALKEGGKVVTI 811
Query: 252 LPPGTPP 258
+PPG PP
Sbjct: 812 VPPGFPP 832
>TC88430 weakly similar to PIR|T10824|T10824 auxin-induced protein (clone
MII-3) - mung bean, partial (41%)
Length = 1450
Score = 285 bits (729), Expect = 2e-77
Identities = 146/307 (47%), Positives = 208/307 (67%), Gaps = 2/307 (0%)
Frame = +2
Query: 12 KAWVYSQYGNIEEILKFDSNVPTPHPKEDQVLIKVVAAALNPVDIKRALGHFKDIDSPLP 71
KAW Y +YG +E+LK + P P P E+Q+L++V AAALNP+D KR + S P
Sbjct: 35 KAWFYEEYGP-KEVLKL-GDFPIPSPLENQLLVQVYAAALNPIDSKRRMRPI--FPSDFP 202
Query: 72 TVPGYDVAGVVVSVGEQVKKFKVGDEVYGDINEI-TLHNPKTIGTLSEYTVAEEKVLAHK 130
VPG D+AGVV+ G VKKF +GDEVYG+I + ++ PK +GTL+++ V EE ++A K
Sbjct: 203 AVPGCDMAGVVIGKGVNVKKFDIGDEVYGNIQDFNSMEKPKQLGTLAQFIVVEENLVARK 382
Query: 131 PSNLSFVEAASLPLAIITAYQGLERVEFSSGKSLLVLGGAGGVGSLVIQLAKHVFEASRI 190
P LSF EAASLPLA+ TA +G + +F G+++ V+GGAGGVG+LV+QLAK +F AS +
Sbjct: 383 PKRLSFEEAASLPLAVQTAIEGFKTGDFKKGETMFVVGGAGGVGTLVLQLAKLMFGASYV 562
Query: 191 AATASTTKLDFLRKLGADLAIDYTKENYEELTEKFDVVYDAVGDSERAVKAANEGGKVVT 250
++ ST KL F+++ GAD +DYTK YE++ EKFD +YD VGD +++ A + G +V
Sbjct: 563 VSSCSTPKLKFVKQFGADKVVDYTKTKYEDIEEKFDFIYDTVGDCKKSFVVAKKDGAIVD 742
Query: 251 IL-PPGTPPAIPFLLTSDGAVLEKLQPYLENGKVKPILDPKSPFPFYQTLEAFSYLNTNR 309
I P A+ LT G +LEKL+PYLE G++K ++DPK + F ++AF Y+ T R
Sbjct: 743 ITWPASHERAVYSSLTVCGEILEKLRPYLERGELKAVIDPKGEYDFENVIDAFGYIETGR 922
Query: 310 AIGKIVI 316
A GK+V+
Sbjct: 923 AWGKVVV 943
>TC86710 similar to GP|15724256|gb|AAL06521.1 AT4g13010/F25G13_100
{Arabidopsis thaliana}, partial (98%)
Length = 1176
Score = 137 bits (344), Expect = 7e-33
Identities = 104/309 (33%), Positives = 148/309 (47%), Gaps = 28/309 (9%)
Frame = +3
Query: 12 KAWVYSQYGNIEEILKFDSNVPTPHPKEDQVLIKVVAAALNPVDIKRALGHFKDIDSP-- 69
+A YS YG LK + VP P PK ++VL+K+ A ++NP+D K G + I P
Sbjct: 60 QALQYSSYGGGVSGLKH-AEVPVPIPKTNEVLLKLEATSINPIDWKIQSGALRAIFLPRK 236
Query: 70 LPTVPGYDVAGVVVSVGEQVKKFKVGDEVYGDINEITLHNPKTIGTLSEYTVAEEKVLAH 129
P +P DVAG VV VG QVK FK GD+V + + G L+E+ VA E + A
Sbjct: 237 FPHIPCTDVAGEVVEVGPQVKDFKSGDKVIAKLTH------QYGGGLAEFAVASESLTAT 398
Query: 130 KPSNLSFVEAASLPLAIITAYQGLERV------EFSSGKSLLVLGGAGGVGSLVIQLAKH 183
+P +S EAA LP+A +TA L + K++LV +GGVGS +QLAK
Sbjct: 399 RPPEVSAAEAAGLPIAGLTARDALTEIGGIKLDGTGEQKNVLVTAASGGVGSFAVQLAK- 575
Query: 184 VFEASRIAATASTTKLDFLRKLGADLAIDYTKENYEELTEKFDVVYDAVGDSERAVKAA- 242
+ + AT +DF++ LGAD +DY L YDAV + +
Sbjct: 576 -LGNNHVTATCGARNIDFVKSLGADEVLDYKTPEGTSLKSPSGKKYDAVIHCTTGIPWST 752
Query: 243 -----NEGGKVVTILPP--------------GTPPAIPFLLTSDGAVLEKLQPYLENGKV 283
+E G VV + P +PF++T LE L +++GK+
Sbjct: 753 FDPNLSEKGVVVDLTPGPSSMLTFALKKTTFSKKRLVPFVVTVKREGLEHLAQLVKDGKL 932
Query: 284 KPILDPKSP 292
K I+D K P
Sbjct: 933 KTIIDSKFP 959
>BE317701 similar to GP|14532287|gb| quinone oxidoreductase-like protein
{Helianthus annuus}, partial (28%)
Length = 269
Score = 122 bits (306), Expect = 2e-28
Identities = 61/88 (69%), Positives = 70/88 (79%)
Frame = +3
Query: 80 GVVVSVGEQVKKFKVGDEVYGDINEITLHNPKTIGTLSEYTVAEEKVLAHKPSNLSFVEA 139
GVVV VG +VK FKVGDEVYGD+NE L P G+L+EYT EEK+LA KP+NL F +A
Sbjct: 3 GVVVKVGSEVKDFKVGDEVYGDVNEKALEGPNQFGSLAEYTAVEEKLLALKPTNLDFAQA 182
Query: 140 ASLPLAIITAYQGLERVEFSSGKSLLVL 167
ASLPLAI TAY+GLER FSSGKS+LVL
Sbjct: 183 ASLPLAIETAYEGLERTGFSSGKSILVL 266
>TC90889 similar to GP|15724256|gb|AAL06521.1 AT4g13010/F25G13_100
{Arabidopsis thaliana}, partial (69%)
Length = 779
Score = 110 bits (276), Expect = 6e-25
Identities = 76/208 (36%), Positives = 105/208 (49%), Gaps = 7/208 (3%)
Frame = +3
Query: 32 VPTPHPKEDQVLIKVVAAALNPVDIKRALGHFKDI-DSPLPTVPGYDVAGVVVSVGEQVK 90
V P PK ++VLIKV A+++NPVD K G + + P P DVAG VV G QVK
Sbjct: 120 VHIPTPKANEVLIKVEASSINPVDWKIQDGLLRYLLPRKFPFTPCTDVAGEVVDFGSQVK 299
Query: 91 KFKVGDEVYGDINEITLHNPKTIGTLSEYTVAEEKVLAHKPSNLSFVEAASLPLA----- 145
FKVGD+V +N + G L+E+ VA E + +PS +S E A LP+A
Sbjct: 300 DFKVGDKVIAKLNN------QDGGGLAEFAVASESLTTLRPSEVSAAEGAGLPVAGLAAH 461
Query: 146 -IITAYQGLERVEFSSGKSLLVLGGAGGVGSLVIQLAKHVFEASRIAATASTTKLDFLRK 204
IT G++ K++LV +GGVG +QLAK + + AT ++ ++
Sbjct: 462 DAITKMAGIKLDRTGEPKNILVTAASGGVGVYAVQLAK--LGNNHVTATCGARNIELIKS 635
Query: 205 LGADLAIDYTKENYEELTEKFDVVYDAV 232
LGAD +DY L YDAV
Sbjct: 636 LGADEVLDYKTPEGASLKSPSGRKYDAV 719
>TC78193 similar to PIR|T05166|T05166 quinone reductase homolog F18E5.200 -
Arabidopsis thaliana, partial (95%)
Length = 1379
Score = 108 bits (271), Expect = 2e-24
Identities = 101/338 (29%), Positives = 162/338 (47%), Gaps = 33/338 (9%)
Frame = +2
Query: 12 KAWVYSQYGNIEEILKFDSNVPTPHPKEDQVLIKVVAAALNPVDIKRALGHFKDIDSPLP 71
KA V ++ G E+L+ V P K+D+VLIKV A ALN D + G + P
Sbjct: 20 KAIVITKPGE-PEVLQLQE-VQDPQIKDDEVLIKVHATALNRADTLQRKGAYPPPQGASP 193
Query: 72 TVPGYDVAGVVVSVGEQVKKFKVGDEVYGDINEITLHNPKTIGTLSEYTVAEEKVLAHKP 131
PG + +G++ VG+ V K+K+GD+V + G + VAE +VL P
Sbjct: 194 -YPGLECSGIIEFVGKNVSKWKIGDQVCALL--------AGGGYAEKVAVAEGQVLP-VP 343
Query: 132 SNLSFVEAASLPLAIITAYQGLERVE-FSSGKSLLVLGGAGGVGSLVIQLAKHVFEASRI 190
+S +AAS P T + + S G++LL+ GG+ G+G+ IQ+AK++ SR+
Sbjct: 344 PGISLKDAASFPEVACTVWSTIFMTSRLSKGETLLIHGGSSGIGTFAIQIAKYL--GSRV 517
Query: 191 AATA-STTKLDFLRKLGADLAIDYTKENY------EELTEKFDVVYDAVGDS--ERAVKA 241
TA S KL F + +GAD+ I+Y E++ E + DV+ D +G S +R + +
Sbjct: 518 FVTAGSEEKLAFCKSIGADVGINYKTEDFVARVKEETGGQGVDVILDCMGASYYQRNLAS 697
Query: 242 ANEGGKV----------------------VTILPPGTPPAIPFLLTSDGAVLEK-LQPYL 278
N G++ +T+ G P A +EK + P +
Sbjct: 698 LNFDGRLFIIGFQGGVSTELDLRALFGKRLTVQAAGLRSRSPENKAVIVAEVEKNVWPAI 877
Query: 279 ENGKVKPILDPKSPFPFYQTLEAFSYLNTNRAIGKIVI 316
GKVKP++ FP + EA + +++ IGKI++
Sbjct: 878 AEGKVKPVV--YKSFPLSEAAEAHRLMESSQHIGKILL 985
>BF650375 similar to GP|15724256|gb AT4g13010/F25G13_100 {Arabidopsis
thaliana}, partial (55%)
Length = 564
Score = 105 bits (263), Expect = 2e-23
Identities = 71/175 (40%), Positives = 99/175 (56%), Gaps = 8/175 (4%)
Frame = +1
Query: 16 YSQYGNIEEILKFDSNVPTPHPKEDQVLIKVVAAALNPVDIKRALGHFKDIDSP--LPTV 73
Y+ YG LK VP P PK ++VLIK+ A ++NP+D K G + + P P +
Sbjct: 25 YNSYGGGASGLKH-VEVPIPTPKTNEVLIKLEAISINPLDWKIQQGVLRAMFFPRKFPHI 201
Query: 74 PGYDVAGVVVSVGEQVKKFKVGDEVYGDINEITLHNPKTIGTLSEYTVAEEKVLAHKPSN 133
P DVAG VV VG +VK FKVGD+V + + G L+E+ VA E + A +PS
Sbjct: 202 PCTDVAGEVVEVGPRVKDFKVGDKVIAKLTN------EYGGGLAEFAVASESLTAARPSE 363
Query: 134 LSFVEAASLPLAIITAYQ------GLERVEFSSGKSLLVLGGAGGVGSLVIQLAK 182
+S EAA LP+A ITA+ G++ K++LV +GGVG+ V+QLAK
Sbjct: 364 VSAAEAAGLPIAGITAHDALTKNGGIKLDGTGEQKNILVTAASGGVGAYVVQLAK 528
>TC79261 similar to PIR|T49047|T49047 quinone reductase-like protein -
Arabidopsis thaliana, partial (95%)
Length = 1239
Score = 100 bits (250), Expect = 6e-22
Identities = 76/243 (31%), Positives = 118/243 (48%), Gaps = 16/243 (6%)
Frame = +3
Query: 35 PHPKEDQ---VLIKVVAAALNPVDIKRALGHFKDIDSPLPTVPGYDVAGVVVSVGEQVKK 91
P P+ D V +++ A +LN + + LG +++ LP +PG D +G+V SVG V
Sbjct: 114 PIPQLDSPTSVRVRIKATSLNFANYLQILGKYQE-KPHLPFIPGSDFSGIVDSVGSNVTN 290
Query: 92 FKVGDEVYGDINEITLHNPKTIGTLSEYTVAEEKVLAHKPSNLSFVEAASLPLAIITAYQ 151
F+VGD V + +G+ ++Y V ++ L P V A +L +A T++
Sbjct: 291 FRVGDPVC---------SFAALGSYAQYLVVDQTQLFRVPEGCDLVAAGALAVAFGTSHV 443
Query: 152 GL-ERVEFSSGKSLLVLGGAGGVGSLVIQLAKHVFEASRIAATASTTKLDFLRKLGADLA 210
GL R + SG+ LLVLG AGGVG +Q+ K A IA K+ L+ +G D
Sbjct: 444 GLVHRAQLKSGQVLLVLGAAGGVGLAAVQIGK-ACGAIVIAVARGAEKVQLLKSMGVDHV 620
Query: 211 IDYTKENYEELTEKF---------DVVYDAVGDS--ERAVKAANEGGKVVTI-LPPGTPP 258
+D EN E ++F DV+YD VG + +++ G ++ I G P
Sbjct: 621 VDLGNENVTESVKEFLKVKRLKGIDVLYDPVGGKLMKESLRLLKWGANILIIGFASGEIP 800
Query: 259 AIP 261
IP
Sbjct: 801 VIP 809
>TC84920 similar to GP|15724256|gb|AAL06521.1 AT4g13010/F25G13_100
{Arabidopsis thaliana}, partial (54%)
Length = 563
Score = 95.9 bits (237), Expect = 2e-20
Identities = 63/154 (40%), Positives = 84/154 (53%), Gaps = 8/154 (5%)
Frame = +2
Query: 37 PKEDQVLIKVVAAALNPVDIKRALGHFKDIDSP--LPTVPGYDVAGVVVSVGEQVKKFKV 94
PK ++VLIK+ A +NP D K G + I P P P DVAG VV +GEQVK FKV
Sbjct: 68 PKTNEVLIKLEAIRINPADWKIQTGVLRAIFFPRKFPHTPCTDVAGEVVEIGEQVKDFKV 247
Query: 95 GDEVYGDINEITLHNPKTIGTLSEYTVAEEKVLAHKPSNLSFVEAASLPLAIITAYQGLE 154
GD+V +N + G L+E+ VA E + A +PS +S EAA LP+A +TA L
Sbjct: 248 GDKVIAKLNNL------YGGGLAEFAVARESLTAARPSEVSAAEAAGLPIAGLTARDALT 409
Query: 155 RV------EFSSGKSLLVLGGAGGVGSLVIQLAK 182
+ K++LV +GGV +QLAK
Sbjct: 410 EIGGVKLDGTGEPKNVLVTAASGGVXVYAVQLAK 511
>TC77119 similar to PIR|D96533|D96533 ARP protein [imported] - Arabidopsis
thaliana, partial (93%)
Length = 2277
Score = 95.5 bits (236), Expect = 2e-20
Identities = 84/338 (24%), Positives = 146/338 (42%), Gaps = 40/338 (11%)
Frame = +1
Query: 19 YGNIEEILKFDSNVPTPHPKEDQVLIKVVAAALNPVDIKRALGHF-----KDIDSPLPTV 73
+ N I++ +P K + VL+K++ A +N D+ + G + K+ + LP
Sbjct: 988 FRNATSIVRAPLRLPV---KPNLVLVKIIYAGVNASDVNFSSGRYFGGNNKETTARLPFD 1158
Query: 74 PGYDVAGVVVSVGEQVKKFKVGDEVYGDINEITLHNPKTIGTLSEYTVAEEKVLAHKPSN 133
G++ G++ +VG+ V KVG T G +E+T+ K P
Sbjct: 1159 AGFEAVGIIAAVGDSVTDLKVGMPCAF----------MTFGGYAEFTMIPSKYALPVPR- 1305
Query: 134 LSFVEAASLPLAIITAYQGLERV-EFSSGKSLLVLGGAGGVGSLVIQLAKHVFEASRIAA 192
E ++ + +TA LE+ + SGK +LV AGG G +QLAK + + +A
Sbjct: 1306 -PDPEGVAMLTSGLTASIALEKAGQMESGKVVLVTAAAGGTGQFAVQLAK-LAGNTVVAT 1479
Query: 193 TASTTKLDFLRKLGADLAIDYTKENYEELTEK-----FDVVYDAVGDS--ERAVKAANEG 245
TK L++LG D IDY E+ + + K D++Y++VG + + A
Sbjct: 1480 CGGGTKAKLLKELGVDRVIDYNSEDIKTVLRKEFPKGIDIIYESVGGDMLKLCLDALAVH 1659
Query: 246 GKVVTI------------LPPGTPPAIPFLLTSDGAV---------------LEKLQPYL 278
G+++ I P P + LL V L++L
Sbjct: 1660 GRLIVIGMISQYQGEHGWTPSKYPGLLEKLLAKSQTVAGFFLVQYSHFYQEHLDRLFDLY 1839
Query: 279 ENGKVKPILDPKSPFPFYQTLEAFSYLNTNRAIGKIVI 316
GK+K +DPK + +A YL++ +++GK+V+
Sbjct: 1840 SKGKLKVAVDPKKFIGLHSVADAVEYLHSGKSVGKVVV 1953
Score = 28.1 bits (61), Expect = 4.7
Identities = 36/118 (30%), Positives = 49/118 (41%), Gaps = 11/118 (9%)
Frame = +1
Query: 156 VEFSSGKSLLVLGGAGGVG-SLVIQLA-KHVFEASRIAATASTTKLDFLRKLGADLAIDY 213
+E +G S LV GGA G+G LV+ LA K VF T +DF + G + A
Sbjct: 94 MEIKAGLSALVTGGASGIGKGLVLALAEKGVF----------ITIVDFSEEKGRETATLV 243
Query: 214 TKEN---YEELTEKFDVVYDAVGDSERAVKAANE------GGKVVTILPPGTPPAIPF 262
K N + L + + R + AA E GG + I+ G IPF
Sbjct: 244 EKINTKFHPNLHHPSVLFVKCDVTNSRDLAAAFEKHVSTFGGLDICIISAGIENPIPF 417
>TC77120 similar to PIR|D96533|D96533 ARP protein [imported] - Arabidopsis
thaliana, partial (93%)
Length = 2203
Score = 94.0 bits (232), Expect = 7e-20
Identities = 82/319 (25%), Positives = 140/319 (43%), Gaps = 40/319 (12%)
Frame = +3
Query: 38 KEDQVLIKVVAAALNPVDIKRALGHF-----KDIDSPLPTVPGYDVAGVVVSVGEQVKKF 92
K + VL+K++ A +N D+ + G + K+ + LP G++ G++ +VG+ V
Sbjct: 1062 KPNHVLVKIIYAGVNASDVNFSSGRYFGGNNKETAARLPFDAGFEAVGIIAAVGDSVTDL 1241
Query: 93 KVGDEVYGDINEITLHNPKTIGTLSEYTVAEEKVLAHKPSNLSFVEAASLPLAIITAYQG 152
KVG T G +E+T+ K P EA ++ + +TA
Sbjct: 1242 KVGMPCAF----------MTFGGYAEFTMIPSKYALPVPR--PDPEAVAMLTSGLTASIA 1385
Query: 153 LERV-EFSSGKSLLVLGGAGGVGSLVIQLAKHVFEASRIAATASTTKLDFLRKLGADLAI 211
LE+ + SGK +LV AGG G +QLAK + + +A K L++LG D I
Sbjct: 1386 LEKAGQMESGKVVLVTAAAGGTGQFAVQLAK-LAGNTVVATCGGGAKAKLLKELGVDRVI 1562
Query: 212 DYTKENYEELTEK-----FDVVYDAVGDS--ERAVKAANEGGKVVTI------------L 252
DY E+ + + +K D++Y++VG + + A G+++ I
Sbjct: 1563 DYHSEDIKTVLKKEFPKGIDIIYESVGGDMLKLCLDALAVHGRLIVIGMISQYQGEKGWT 1742
Query: 253 PPGTPPAIPFLLTSDGAV---------------LEKLQPYLENGKVKPILDPKSPFPFYQ 297
P P LL+ AV L++L GK+K +DPK +
Sbjct: 1743 PSKYPGLCEKLLSKSQAVAGFFLVQYSHMWQEHLDRLFDLYSQGKLKVAVDPKKFIGLHS 1922
Query: 298 TLEAFSYLNTNRAIGKIVI 316
+A YL++ ++ GK+V+
Sbjct: 1923 VADAVEYLHSGKSAGKVVV 1979
Score = 31.6 bits (70), Expect = 0.43
Identities = 26/68 (38%), Positives = 34/68 (49%), Gaps = 2/68 (2%)
Frame = +3
Query: 152 GLERVEFSSGKSLLVLGGAGGVG-SLVIQLA-KHVFEASRIAATASTTKLDFLRKLGADL 209
G +R+E +G S LV GGA G+G LV+ LA K VF T +DF + G +
Sbjct: 108 GTKRMEIKAGLSALVTGGASGIGKGLVLALAEKGVF----------ITIVDFSEEKGRET 257
Query: 210 AIDYTKEN 217
A K N
Sbjct: 258 ATLVEKIN 281
>AW127671 weakly similar to GP|15724256|gb AT4g13010/F25G13_100 {Arabidopsis
thaliana}, partial (48%)
Length = 493
Score = 88.2 bits (217), Expect = 4e-18
Identities = 61/147 (41%), Positives = 82/147 (55%), Gaps = 3/147 (2%)
Frame = +2
Query: 12 KAWVYSQYGNIEEILKFDSNVPTPHPKEDQVLIKVVAAALNPVDIK---RALGHFKDIDS 68
+A Y+ YG LK VP P P +D+VLIK+ AA++NP D K R L F +
Sbjct: 26 RAVQYNAYGGGSTGLKH-VEVPIPSPSKDEVLIKLEAASINPFDWKVQKRMLWPF--LPP 196
Query: 69 PLPTVPGYDVAGVVVSVGEQVKKFKVGDEVYGDINEITLHNPKTIGTLSEYTVAEEKVLA 128
P +P DVAG V+ VG+ VKKFK GD+V G L +P + G L+E+ V +E V A
Sbjct: 197 KFPYIPCTDVAGEVMMVGKGVKKFKTGDKVVG------LVSPFSGGGLAEFAVVKESVTA 358
Query: 129 HKPSNLSFVEAASLPLAIITAYQGLER 155
P +S E LP+A +TA Q L +
Sbjct: 359 SIPQEISASECVGLPVAGLTALQALTK 439
>TC78194 similar to PIR|T05166|T05166 quinone reductase homolog F18E5.200 -
Arabidopsis thaliana, partial (60%)
Length = 709
Score = 85.9 bits (211), Expect = 2e-17
Identities = 62/197 (31%), Positives = 98/197 (49%), Gaps = 2/197 (1%)
Frame = +1
Query: 24 EILKFDSNVPTPHPKEDQVLIKVVAAALNPVDIKRALGHFKDIDSPLPTVPGYDVAGVVV 83
E+L+ V P PK+++VLI+V A ALN D + G F I PG + +G +V
Sbjct: 157 EVLQLQE-VQDPQPKDNEVLIRVHATALNSADTVQRKG-FYPIPQGASPYPGLECSGTIV 330
Query: 84 SVGEQVKKFKVGDEVYGDINEITLHNPKTIGTLSEYTVAEEKVLAHKPSNLSFVEAASLP 143
VG V K+K+GD+V + G +E E + P +S +AAS
Sbjct: 331 CVGINVSKWKIGDQVCALLAG---------GGYAEKVAVPEGQVLPVPPGISLKDAASFT 483
Query: 144 LAIITAYQGLERVE-FSSGKSLLVLGGAGGVGSLVIQLAKHVFEASRIAATA-STTKLDF 201
T + + S G++LLV GG+ G+G+ I +AK ++ S++ TA S KL F
Sbjct: 484 EVACTVWSTIFMTSRLSKGETLLVHGGSSGIGTFAIXIAK--YQGSKVFVTAGSEEKLAF 657
Query: 202 LRKLGADLAIDYTKENY 218
+ +GAD+ +Y ++
Sbjct: 658 CKSIGADVGXNYXTXDF 708
>TC87521 similar to SP|Q9ZRF1|MTD_FRAAN Probable mannitol dehydrogenase (EC
1.1.1.255) (NAD-dependent mannitol dehydrogenase).
[Strawberry], partial (98%)
Length = 1433
Score = 73.9 bits (180), Expect = 7e-14
Identities = 64/226 (28%), Positives = 96/226 (42%), Gaps = 30/226 (13%)
Frame = +2
Query: 68 SPLPTVPGYDVAGVVVSVGEQVKKFKVGDEV-YGDIN---------EITLHN--PK---- 111
S P VPG+++AG+V VG +V+KFK+GD+V G + E L N PK
Sbjct: 257 STYPLVPGHEIAGIVTEVGSKVEKFKIGDKVGVGCLVDSCRACQNCEENLENYCPKQTNT 436
Query: 112 ----------TIGTLSEYTVAEEKVLAHKPSNLSFVEAASLPLAIITAYQGLERVEFSSG 161
T G S+ VA+E + H P L AA L A IT Y L
Sbjct: 437 YSAKYSDGSITYGGYSDSMVADEHFIVHIPDGLPLESAAPLLCAGITVYSPLRYFGLDKP 616
Query: 162 KSLLVLGGAGGVGSLVIQLAKHVFEASRIAATASTTKLDFLRKLGAD-LAIDYTKENYEE 220
+ + G GG+G L ++ AK + +T+ + + + LGAD I + ++ +
Sbjct: 617 GMNIGIVGLGGLGHLGVKFAKAFGANVTVISTSPNKEKEAIENLGADSFLISHDQDKMQA 796
Query: 221 LTEKFDVVYDAVGDSE---RAVKAANEGGKVVTILPPGTPPAIPFL 263
D + D V V GK+V + P PP +P +
Sbjct: 797 AMGTLDGIIDTVSADHPLLPLVGLLKYHGKLVMVGAPDKPPELPHI 934
>BI263584 similar to SP|Q9P6C8|ADH1 Alcohol dehydrogenase I (EC 1.1.1.1).
{Neurospora crassa}, partial (57%)
Length = 682
Score = 68.9 bits (167), Expect = 2e-12
Identities = 52/199 (26%), Positives = 87/199 (43%), Gaps = 19/199 (9%)
Frame = +2
Query: 3 STPSIPSHTKAWVYSQYGNIEEILKFDSNVPTPHPKEDQVLIKVVAAALNPVDIKRALGH 62
+ P IP A V+ + E +P P D+VL+ V + + D+ G
Sbjct: 65 AAPQIPEKQWAQVFEKTAGPIEY----KQIPVQKPGPDEVLVNVKFSGVCHTDLHAWQGD 232
Query: 63 FKDIDSPLPTVPGYDVAGVVVSVGEQVKKFKVGDEV-------------------YGDIN 103
+ +D+ LP V G++ AGVVV+ G+ VK K+G++V
Sbjct: 233 WP-LDTKLPLVGGHEGAGVVVARGDLVKDVKIGEKVGIKWLNGSCLSCSYCQNADESLCA 409
Query: 104 EITLHNPKTIGTLSEYTVAEEKVLAHKPSNLSFVEAASLPLAIITAYQGLERVEFSSGKS 163
E L G+ +Y +A+ +A P + + A IT Y+GL+ +G+S
Sbjct: 410 EALLSGYTVDGSFQQYAIAKAIHVARIPEECDLESISPILCAGITVYKGLKESGVKAGQS 589
Query: 164 LLVLGGAGGVGSLVIQLAK 182
+ ++G GG+GS+ Q K
Sbjct: 590 IAIVGAGGGLGSIAXQYCK 646
>TC79080 similar to GP|10177043|dbj|BAB10455. alcohol dehydrogenase-like
protein {Arabidopsis thaliana}, partial (96%)
Length = 1524
Score = 67.8 bits (164), Expect = 5e-12
Identities = 76/301 (25%), Positives = 128/301 (42%), Gaps = 56/301 (18%)
Frame = +3
Query: 7 IPSHTKAWVYSQYGNIEEILKFDSNVPTPHPKEDQVLIKVVAAALNPVDIKRALGHFKDI 66
+PS+ + V+ + I +F P PK ++LIK + D+ G +I
Sbjct: 219 LPSYMRGAVFREPNKPLTIEEFH----IPRPKAGELLIKTKGCGVCHSDLHVMKG---EI 377
Query: 67 DSPLPTVPGYDVAGVVVSVGEQ-----VKKFKVGDEVYG-----------------DINE 104
P V G+++ G VV G+ +++ +G V G D+ E
Sbjct: 378 PFSSPCVVGHEITGEVVEHGQHTDSKTIERLPIGSRVVGAFIMPCGNCSYCSKGHDDLCE 557
Query: 105 I---------TLHNPKT---------------IGTLSEYTVAEEKVLAHKPSNLSFVEAA 140
TL++ +T +G L+EY V LA P+++ + E+A
Sbjct: 558 AFFAYNRAKGTLYDGETRLFLRGSGNPIYMYSMGGLAEYCVVPANALAVLPNSMPYTESA 737
Query: 141 SLPLAIITAYQGLERV-EFSSGKSLLVLGGAGGVGSLVIQLAKHVFEASRIAATASTTKL 199
L A+ TAY + E G ++ V+G GGVGS +Q+A+ + IA KL
Sbjct: 738 ILGCAVFTAYGAMAHAAEVRPGDTVAVIG-TGGVGSSCLQIARAFGASDIIAVDVQDEKL 914
Query: 200 DFLRKLGADLAIDYTKEN-YEELTE-----KFDVVYDAVGDSE---RAVKAANEGGKVVT 250
+ + LGA I+ KE+ E++ E DV +A+G + + ++ +GGK V
Sbjct: 915 EKAKTLGATHTINSAKEDPIEKILEITGGKGVDVAVEALGRPQTFAQCTQSVKDGGKAVM 1094
Query: 251 I 251
I
Sbjct: 1095I 1097
>TC88383 similar to GP|21592515|gb|AAM64465.1 nuclear receptor binding
factor-like protein {Arabidopsis thaliana}, partial
(90%)
Length = 1313
Score = 62.0 bits (149), Expect(2) = 3e-11
Identities = 45/153 (29%), Positives = 73/153 (47%)
Frame = +3
Query: 2 ASTPSIPSHTKAWVYSQYGNIEEILKFDSNVPTPHPKEDQVLIKVVAAALNPVDIKRALG 61
A + ++ +KA +Y +G + + K ++P KE+ + +K++AA +NP DI R G
Sbjct: 96 AFSSAVSPPSKAIIYESHGQPDAVTKL-VDIPATEVKENDLCVKMLAAPINPSDINRIQG 272
Query: 62 HFKDIDSPLPTVPGYDVAGVVVSVGEQVKKFKVGDEVYGDINEITLHNPKTIGTLSEYTV 121
+ P P V GY+ G V SVG V F GD V + +P + GT Y V
Sbjct: 273 VYPVRPEP-PAVGGYEGVGEVHSVGSAVTCFSPGDWV--------IPSPPSFGTWQTYIV 425
Query: 122 AEEKVLAHKPSNLSFVEAASLPLAIITAYQGLE 154
++ V + AA++ + +TA LE
Sbjct: 426 KDQNVWHKVNKGVPMEYAATITVNPLTALLMLE 524
Score = 23.1 bits (48), Expect(2) = 3e-11
Identities = 19/54 (35%), Positives = 25/54 (46%), Gaps = 4/54 (7%)
Frame = +1
Query: 159 SSGKSLLVLGGAGGVGSLVIQLAK----HVFEASRIAATASTTKLDFLRKLGAD 208
+SG +++ G VG VIQLAK H R K + L+ LGAD
Sbjct: 541 NSGDAIVQNGATSMVGQCVIQLAKSRGIHNINIIRDRPGVGEVK-ERLKDLGAD 699
>TC85885 similar to SP|Q9ZRF1|MTD_FRAAN Probable mannitol dehydrogenase (EC
1.1.1.255) (NAD-dependent mannitol dehydrogenase).
[Strawberry], partial (97%)
Length = 1330
Score = 60.1 bits (144), Expect = 1e-09
Identities = 50/196 (25%), Positives = 82/196 (41%), Gaps = 26/196 (13%)
Frame = +3
Query: 39 EDQVLIKVVAAALNPVDIKRALGHFKDIDSPLPTVPGYDVAGVVVSVGEQVKKFKVGDEV 98
E V KV+ + D+ + + S P VPG+++AG+V VG +V+KFKVGD+V
Sbjct: 177 EKDVAFKVLYCGICHTDLHMMKNEWGN--SIYPLVPGHELAGIVTEVGSKVEKFKVGDKV 350
Query: 99 ---------------------YGDINEITL-----HNPKTIGTLSEYTVAEEKVLAHKPS 132
Y +T T G S+ VA+E + P
Sbjct: 351 GVGYMVDSCRSCENCAEDLENYCPQQTVTCGAKYRDGSVTYGGYSDSMVADEHFVIRIPD 530
Query: 133 NLSFVEAASLPLAIITAYQGLERVEFSSGKSLLVLGGAGGVGSLVIQLAKHVFEASRIAA 192
+L A L A +T Y L + + + G GG+G + ++ AK + +
Sbjct: 531 SLPLDVAGPLLCAGVTVYSPLRHFQLDKPGMNIGVVGLGGLGHMAVKFAKAFGANVTVIS 710
Query: 193 TASTTKLDFLRKLGAD 208
T+ + + + + LGAD
Sbjct: 711 TSPSKEKEAIEHLGAD 758
>TC80850 homologue to SP|O82515|MTD_MEDSA Probable mannitol dehydrogenase
(EC 1.1.1.255) (NAD-dependent mannitol dehydrogenase).
[Alfalfa], partial (61%)
Length = 710
Score = 56.6 bits (135), Expect = 1e-08
Identities = 42/137 (30%), Positives = 61/137 (43%), Gaps = 25/137 (18%)
Frame = +1
Query: 71 PTVPGYDVAGVVVSVGEQVKKFKVGDEV-YGDINE--------------------ITLHN 109
P VPG+++ GVV VG VKKF+VGD V G I E T ++
Sbjct: 250 PVVPGHEIVGVVTKVGINVKKFRVGDNVGVGVIVESCQTCENCNQDLEQYCPKLVFTYNS 429
Query: 110 P----KTIGTLSEYTVAEEKVLAHKPSNLSFVEAASLPLAIITAYQGLERVEFSSGKSLL 165
P +T G S++ V ++ + P NL A L A IT Y ++ + L
Sbjct: 430 PYKGTRTHGGYSDFVVVHQRYVVQFPDNLPLDAGAPLLCAGITVYSPMKYYGMTEPGKHL 609
Query: 166 VLGGAGGVGSLVIQLAK 182
+ G GG+G + I+ K
Sbjct: 610 GVAGLGGLGHVAIKFGK 660
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.315 0.135 0.383
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,359,340
Number of Sequences: 36976
Number of extensions: 106414
Number of successful extensions: 535
Number of sequences better than 10.0: 65
Number of HSP's better than 10.0 without gapping: 500
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 510
length of query: 320
length of database: 9,014,727
effective HSP length: 96
effective length of query: 224
effective length of database: 5,465,031
effective search space: 1224166944
effective search space used: 1224166944
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 58 (26.9 bits)
Medicago: description of AC139600.7


