

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC139600.2 + phase: 0
(98 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
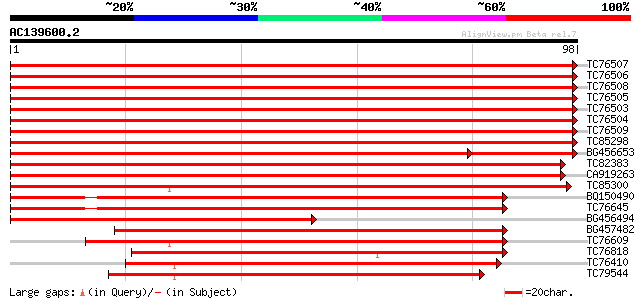
Score E
Sequences producing significant alignments: (bits) Value
TC76507 homologue to GP|506629|gb|AAA50172.1|| photosystem II ty... 194 4e-51
TC76506 homologue to GP|506629|gb|AAA50172.1|| photosystem II ty... 194 4e-51
TC76508 GP|3293555|gb|AAC25775.1| chlorophyll a/b binding protei... 194 5e-51
TC76505 homologue to GP|9587207|gb|AAF89207.1| LHCII type I chlo... 192 2e-50
TC76503 homologue to GP|506629|gb|AAA50172.1|| photosystem II ty... 190 7e-50
TC76504 homologue to GP|506629|gb|AAA50172.1|| photosystem II ty... 190 7e-50
TC76509 homologue to PIR|S29904|S29904 chlorophyll a/b-binding p... 180 7e-47
TC85298 homologue to SP|P27520|CB23_PEA Chlorophyll A-B binding ... 175 3e-45
BG456653 homologue to GP|3293555|gb| chlorophyll a/b binding pro... 155 2e-39
TC82383 similar to GP|15430554|dbj|BAB64412. light-harvesting ch... 147 9e-37
CA919263 similar to GP|12658406|gb light-harvesting complex II p... 142 2e-35
TC85300 homologue to PIR|S33775|S33775 chlorophyll a/b-binding p... 139 1e-34
BQ150490 similar to PIR|T04049|T040 chlorophyll a/b-binding prot... 107 6e-25
TC76645 similar to PIR|S16294|S16294 chlorophyll a/b-binding pro... 107 6e-25
BG456494 homologue to GP|2645999|gb| chlorophyll a/b binding pro... 106 1e-24
BG457482 similar to PIR|T04049|T04 chlorophyll a/b-binding prote... 100 2e-22
TC76609 homologue to PIR|T06411|T06411 probable chlorophyll a/b-... 91 1e-19
TC76818 homologue to GP|13265501|gb|AAG40043.2 AT3g54890 {Arabid... 78 6e-16
TC76410 similar to GP|4689382|gb|AAD27878.1| chlorophyll a/b bin... 77 8e-16
TC79544 similar to PIR|F96510|F96510 light-harvesting complex pr... 74 7e-15
>TC76507 homologue to GP|506629|gb|AAA50172.1|| photosystem II type I
chlorophyll a/b-binding protein {Glycine max}, complete
Length = 1029
Score = 194 bits (494), Expect = 4e-51
Identities = 94/98 (95%), Positives = 96/98 (97%)
Frame = +1
Query: 1 MGAVEGYRIADGPLGEVVDPLYPGGSFDPLGLADDPEAFAELKVKELKNGRLAMFSMFGF 60
MGAVEGYRIA GPLGEVVDPLYPGGSFDPLGLA+DPEAFAELKVKELKNGRLAMFSMFGF
Sbjct: 583 MGAVEGYRIAGGPLGEVVDPLYPGGSFDPLGLAEDPEAFAELKVKELKNGRLAMFSMFGF 762
Query: 61 FVQAIVTEKGPLENLADHIADPVNSNAWAFATNFVPGK 98
FVQAIVT KGPLENLADHIADPVN+NAWAFATNFVPGK
Sbjct: 763 FVQAIVTGKGPLENLADHIADPVNNNAWAFATNFVPGK 876
>TC76506 homologue to GP|506629|gb|AAA50172.1|| photosystem II type I
chlorophyll a/b-binding protein {Glycine max}, complete
Length = 1057
Score = 194 bits (494), Expect = 4e-51
Identities = 94/98 (95%), Positives = 95/98 (96%)
Frame = +1
Query: 1 MGAVEGYRIADGPLGEVVDPLYPGGSFDPLGLADDPEAFAELKVKELKNGRLAMFSMFGF 60
MGAVEGYRIA GPLGEVVDPLYPGGSFDPLGLADDPEAFAELKVKELKNGRLAMFSMFGF
Sbjct: 562 MGAVEGYRIAGGPLGEVVDPLYPGGSFDPLGLADDPEAFAELKVKELKNGRLAMFSMFGF 741
Query: 61 FVQAIVTEKGPLENLADHIADPVNSNAWAFATNFVPGK 98
FVQAIVT KGPLENLADHI DPVN+NAWAFATNFVPGK
Sbjct: 742 FVQAIVTGKGPLENLADHITDPVNNNAWAFATNFVPGK 855
>TC76508 GP|3293555|gb|AAC25775.1| chlorophyll a/b binding protein {Medicago
sativa}, complete
Length = 1190
Score = 194 bits (493), Expect = 5e-51
Identities = 93/98 (94%), Positives = 96/98 (97%)
Frame = +2
Query: 1 MGAVEGYRIADGPLGEVVDPLYPGGSFDPLGLADDPEAFAELKVKELKNGRLAMFSMFGF 60
MGAVEGYRIA GPLGEVVDPLYPGGSFDPLGLADDPEAFAELKVKELKNGRLAMFSMFGF
Sbjct: 581 MGAVEGYRIAGGPLGEVVDPLYPGGSFDPLGLADDPEAFAELKVKELKNGRLAMFSMFGF 760
Query: 61 FVQAIVTEKGPLENLADHIADPVNSNAWAFATNFVPGK 98
FVQAIVT KGPLENLADH+ADPVN+NAWA+ATNFVPGK
Sbjct: 761 FVQAIVTGKGPLENLADHLADPVNNNAWAYATNFVPGK 874
>TC76505 homologue to GP|9587207|gb|AAF89207.1| LHCII type I chlorophyll
a/b-binding protein {Vigna radiata}, complete
Length = 1082
Score = 192 bits (487), Expect = 2e-50
Identities = 91/98 (92%), Positives = 95/98 (96%)
Frame = +1
Query: 1 MGAVEGYRIADGPLGEVVDPLYPGGSFDPLGLADDPEAFAELKVKELKNGRLAMFSMFGF 60
MGAVEGYRIA GPLGEVVDPLYPGGSFDPLGLADDPEAFAELKVKELKNGRLAMFSMFGF
Sbjct: 565 MGAVEGYRIAGGPLGEVVDPLYPGGSFDPLGLADDPEAFAELKVKELKNGRLAMFSMFGF 744
Query: 61 FVQAIVTEKGPLENLADHIADPVNSNAWAFATNFVPGK 98
FVQAIVT KGP+ENLADH+ADPVN+NAWA+ATNF PGK
Sbjct: 745 FVQAIVTGKGPIENLADHLADPVNNNAWAYATNFAPGK 858
>TC76503 homologue to GP|506629|gb|AAA50172.1|| photosystem II type I
chlorophyll a/b-binding protein {Glycine max}, partial
(98%)
Length = 1148
Score = 190 bits (483), Expect = 7e-50
Identities = 90/98 (91%), Positives = 95/98 (96%)
Frame = +1
Query: 1 MGAVEGYRIADGPLGEVVDPLYPGGSFDPLGLADDPEAFAELKVKELKNGRLAMFSMFGF 60
MGAVEGYRIA GPLGEVVDPLYPGGSFDPLGLA+DPEAFAELKVKELKNGRLAMFSMFGF
Sbjct: 556 MGAVEGYRIAGGPLGEVVDPLYPGGSFDPLGLAEDPEAFAELKVKELKNGRLAMFSMFGF 735
Query: 61 FVQAIVTEKGPLENLADHIADPVNSNAWAFATNFVPGK 98
FVQAIVT KGP+ENLADH+ADPVN+NAWA+ATNF PGK
Sbjct: 736 FVQAIVTGKGPIENLADHLADPVNNNAWAYATNFAPGK 849
>TC76504 homologue to GP|506629|gb|AAA50172.1|| photosystem II type I
chlorophyll a/b-binding protein {Glycine max}, complete
Length = 1134
Score = 190 bits (483), Expect = 7e-50
Identities = 90/98 (91%), Positives = 95/98 (96%)
Frame = +1
Query: 1 MGAVEGYRIADGPLGEVVDPLYPGGSFDPLGLADDPEAFAELKVKELKNGRLAMFSMFGF 60
MGAVEGYRIA GPLGEVVDPLYPGGSFDPLGLA+DPEAFAELKVKELKNGRLAMFSMFGF
Sbjct: 547 MGAVEGYRIAGGPLGEVVDPLYPGGSFDPLGLAEDPEAFAELKVKELKNGRLAMFSMFGF 726
Query: 61 FVQAIVTEKGPLENLADHIADPVNSNAWAFATNFVPGK 98
FVQAIVT KGP+ENLADH+ADPVN+NAWA+ATNF PGK
Sbjct: 727 FVQAIVTGKGPIENLADHLADPVNNNAWAYATNFAPGK 840
>TC76509 homologue to PIR|S29904|S29904 chlorophyll a/b-binding protein -
English ivy (fragment), partial (53%)
Length = 481
Score = 180 bits (457), Expect = 7e-47
Identities = 86/98 (87%), Positives = 91/98 (92%)
Frame = +3
Query: 1 MGAVEGYRIADGPLGEVVDPLYPGGSFDPLGLADDPEAFAELKVKELKNGRLAMFSMFGF 60
MGAV RI GPLGEVVDPLYPGGSFDPLGLA+DPEAFAELKVKELKNGRLAMFSMFGF
Sbjct: 21 MGAVRVXRIXGGPLGEVVDPLYPGGSFDPLGLAEDPEAFAELKVKELKNGRLAMFSMFGF 200
Query: 61 FVQAIVTEKGPLENLADHIADPVNSNAWAFATNFVPGK 98
FVQAIVT KGP+ENLADH+ADPVN+NAWA+ATNF PGK
Sbjct: 201 FVQAIVTGKGPIENLADHLADPVNNNAWAYATNFAPGK 314
>TC85298 homologue to SP|P27520|CB23_PEA Chlorophyll A-B binding protein 215
chloroplast precursor (LHCII type II CAB-215) (LHCP).,
complete
Length = 1099
Score = 175 bits (443), Expect = 3e-45
Identities = 81/98 (82%), Positives = 91/98 (92%)
Frame = +2
Query: 1 MGAVEGYRIADGPLGEVVDPLYPGGSFDPLGLADDPEAFAELKVKELKNGRLAMFSMFGF 60
MG VEGYR+ GPLGE +DPLYPGG+FDPLGLADDP+AFAELKVKELKNGRLAMFSMFGF
Sbjct: 560 MGFVEGYRVGGGPLGEGLDPLYPGGAFDPLGLADDPDAFAELKVKELKNGRLAMFSMFGF 739
Query: 61 FVQAIVTEKGPLENLADHIADPVNSNAWAFATNFVPGK 98
FVQAIVT KGP++NL DH+ADPV +NAWA+ATNFVPG+
Sbjct: 740 FVQAIVTGKGPVQNLYDHVADPVANNAWAYATNFVPGQ 853
>BG456653 homologue to GP|3293555|gb| chlorophyll a/b binding protein
{Medicago sativa}, partial (55%)
Length = 511
Score = 155 bits (392), Expect = 2e-39
Identities = 75/80 (93%), Positives = 78/80 (96%)
Frame = -1
Query: 1 MGAVEGYRIADGPLGEVVDPLYPGGSFDPLGLADDPEAFAELKVKELKNGRLAMFSMFGF 60
MGAVEGYRIA GPLGEVVDPLYPGGSFDPLGLA+DPEAFAELKVKELKNGRLAMFSMFGF
Sbjct: 307 MGAVEGYRIAGGPLGEVVDPLYPGGSFDPLGLAEDPEAFAELKVKELKNGRLAMFSMFGF 128
Query: 61 FVQAIVTEKGPLENLADHIA 80
FVQAIVT KGP+ENLADH+A
Sbjct: 127 FVQAIVTGKGPIENLADHLA 68
Score = 41.2 bits (95), Expect = 7e-05
Identities = 16/19 (84%), Positives = 18/19 (94%)
Frame = +3
Query: 80 ADPVNSNAWAFATNFVPGK 98
ADPVN+NAWA+ATNF PGK
Sbjct: 9 ADPVNNNAWAYATNFAPGK 65
>TC82383 similar to GP|15430554|dbj|BAB64412. light-harvesting
chlorophyll-a/b binding protein LhcII-1.3 {Chlamydomonas
reinhardtii}, partial (62%)
Length = 615
Score = 147 bits (370), Expect = 9e-37
Identities = 69/96 (71%), Positives = 81/96 (83%)
Frame = +2
Query: 1 MGAVEGYRIADGPLGEVVDPLYPGGSFDPLGLADDPEAFAELKVKELKNGRLAMFSMFGF 60
MG +EGYR+ GP GE +DPL+PG +FDPLGLADDP+ FAELKVKE+KNGRLAMFSMFGF
Sbjct: 197 MGLIEGYRVNGGPAGEGLDPLHPGEAFDPLGLADDPDTFAELKVKEIKNGRLAMFSMFGF 376
Query: 61 FVQAIVTEKGPLENLADHIADPVNSNAWAFATNFVP 96
FVQAIVT KGP+ NL DH+A+P +N +A AT FVP
Sbjct: 377 FVQAIVTGKGPIANLNDHLANPSVNNGFASATKFVP 484
>CA919263 similar to GP|12658406|gb light-harvesting complex II protein
precursor {Chlamydomonas reinhardtii}, partial (57%)
Length = 592
Score = 142 bits (358), Expect = 2e-35
Identities = 66/96 (68%), Positives = 75/96 (77%)
Frame = -3
Query: 1 MGAVEGYRIADGPLGEVVDPLYPGGSFDPLGLADDPEAFAELKVKELKNGRLAMFSMFGF 60
MG E YR + +PLYPGG FDPLGLADDP+ AELKVKELKNGRLAMFSMFGF
Sbjct: 416 MGLAEAYRANGADFIDAAEPLYPGGPFDPLGLADDPDTLAELKVKELKNGRLAMFSMFGF 237
Query: 61 FVQAIVTEKGPLENLADHIADPVNSNAWAFATNFVP 96
F+QAIVT KGP++NL DH+ADP +NAWA+AT F P
Sbjct: 236 FIQAIVTGKGPIQNLNDHLADPSVANAWAYATKFTP 129
>TC85300 homologue to PIR|S33775|S33775 chlorophyll a/b-binding protein -
garden pea, partial (96%)
Length = 1163
Score = 139 bits (351), Expect = 1e-34
Identities = 72/98 (73%), Positives = 78/98 (79%), Gaps = 1/98 (1%)
Frame = +3
Query: 1 MGAVEGYRIADGPLGEVVDPLYPGGS-FDPLGLADDPEAFAELKVKELKNGRLAMFSMFG 59
MG VEG+RI P + LYPGG FDPLGLADDP FAELKVKE+KNGRLAMFSMFG
Sbjct: 657 MGLVEGFRINGLPGVGEGNNLYPGGQYFDPLGLADDPVTFAELKVKEIKNGRLAMFSMFG 836
Query: 60 FFVQAIVTEKGPLENLADHIADPVNSNAWAFATNFVPG 97
FFVQAIVT KGPLENL DHI +PV +NAW +AT FVPG
Sbjct: 837 FFVQAIVTGKGPLENLLDHIDNPVANNAWVYATKFVPG 950
Score = 33.1 bits (74), Expect = 0.018
Identities = 17/39 (43%), Positives = 24/39 (60%), Gaps = 1/39 (2%)
Frame = +3
Query: 22 YPGG-SFDPLGLADDPEAFAELKVKELKNGRLAMFSMFG 59
+PG +D GL+ DPEAFA+ + E+ +GR AM G
Sbjct: 369 FPGDYGWDTAGLSADPEAFAKNRALEVIHGRWAMLGALG 485
>BQ150490 similar to PIR|T04049|T040 chlorophyll a/b-binding protein CP26
[imported] - Arabidopsis thaliana, partial (35%)
Length = 727
Score = 107 bits (268), Expect = 6e-25
Identities = 52/86 (60%), Positives = 66/86 (76%)
Frame = +3
Query: 1 MGAVEGYRIADGPLGEVVDPLYPGGSFDPLGLADDPEAFAELKVKELKNGRLAMFSMFGF 60
+G E YRI +G E+ D +PGG FDPLGLA+DP+ A LKVKE+KNGRLAMF+M GF
Sbjct: 33 LGGAEYYRITNGL--ELEDKFHPGGPFDPLGLANDPDQAAILKVKEIKNGRLAMFAMLGF 206
Query: 61 FVQAIVTEKGPLENLADHIADPVNSN 86
F+QA VT +GP+ENLA H++DP +N
Sbjct: 207 FIQAYVTGEGPVENLAKHLSDPFGNN 284
>TC76645 similar to PIR|S16294|S16294 chlorophyll a/b-binding protein type I
precursor - tomato, partial (89%)
Length = 1071
Score = 107 bits (268), Expect = 6e-25
Identities = 52/86 (60%), Positives = 66/86 (76%)
Frame = +3
Query: 1 MGAVEGYRIADGPLGEVVDPLYPGGSFDPLGLADDPEAFAELKVKELKNGRLAMFSMFGF 60
+G E YRI +G E+ D +PGG FDPLGLA+DP+ A LKVKE+KNGRLAMF+M GF
Sbjct: 651 LGGAEYYRITNGL--ELEDKFHPGGPFDPLGLANDPDQAAILKVKEIKNGRLAMFAMLGF 824
Query: 61 FVQAIVTEKGPLENLADHIADPVNSN 86
F+QA VT +GP+ENLA H++DP +N
Sbjct: 825 FIQAYVTGEGPVENLAKHLSDPFGNN 902
>BG456494 homologue to GP|2645999|gb| chlorophyll a/b binding protein of
LHCII type I precursor {Panax ginseng}, partial (45%)
Length = 535
Score = 106 bits (265), Expect = 1e-24
Identities = 51/53 (96%), Positives = 52/53 (97%)
Frame = -1
Query: 1 MGAVEGYRIADGPLGEVVDPLYPGGSFDPLGLADDPEAFAELKVKELKNGRLA 53
MGAVEGYRIA GPLGEVVDPLYPGGSFDPLGLA+DPEAFAELKVKELKNGRLA
Sbjct: 331 MGAVEGYRIAGGPLGEVVDPLYPGGSFDPLGLAEDPEAFAELKVKELKNGRLA 173
>BG457482 similar to PIR|T04049|T04 chlorophyll a/b-binding protein CP26
[imported] - Arabidopsis thaliana, partial (37%)
Length = 570
Score = 99.8 bits (247), Expect = 2e-22
Identities = 45/68 (66%), Positives = 56/68 (82%)
Frame = +1
Query: 19 DPLYPGGSFDPLGLADDPEAFAELKVKELKNGRLAMFSMFGFFVQAIVTEKGPLENLADH 78
D +PGG FDPLGLA+DP+ A LKVKE+KNGRLAMF+M GFF+QA VT +GP+ENLA H
Sbjct: 202 DKFHPGGPFDPLGLANDPDQAAILKVKEIKNGRLAMFAMLGFFIQAYVTGEGPVENLAKH 381
Query: 79 IADPVNSN 86
++DP +N
Sbjct: 382 LSDPFGNN 405
>TC76609 homologue to PIR|T06411|T06411 probable chlorophyll a/b-binding
protein type III precursor - garden pea chloroplast,
complete
Length = 1500
Score = 90.5 bits (223), Expect = 1e-19
Identities = 42/74 (56%), Positives = 56/74 (74%), Gaps = 1/74 (1%)
Frame = +2
Query: 14 LGEVVDPLYPGGS-FDPLGLADDPEAFAELKVKELKNGRLAMFSMFGFFVQAIVTEKGPL 72
LG +P YPGG F+PLG D ++ ELK+KE+KNGRLAM ++ G+F+QA+VT GP
Sbjct: 722 LGGSGEPAYPGGPIFNPLGFGKDEKSLKELKLKEVKNGRLAMLAILGYFIQALVTGVGPY 901
Query: 73 ENLADHIADPVNSN 86
+NL DH+ADPVN+N
Sbjct: 902 QNLLDHLADPVNNN 943
>TC76818 homologue to GP|13265501|gb|AAG40043.2 AT3g54890 {Arabidopsis
thaliana}, partial (84%)
Length = 1010
Score = 77.8 bits (190), Expect = 6e-16
Identities = 37/66 (56%), Positives = 47/66 (71%), Gaps = 1/66 (1%)
Frame = +1
Query: 22 YPGGSFDPLGLADDPEAFAELKVKELKNGRLAMFSMFGFFV-QAIVTEKGPLENLADHIA 80
YPGG+FDPLG + DP+ F E K+KE+KNGRLA+ + G V Q GPLENLA H+A
Sbjct: 601 YPGGAFDPLGYSKDPKKFHEYKIKEVKNGRLALLAFVGIVVQQTAYPGTGPLENLATHLA 780
Query: 81 DPVNSN 86
DP ++N
Sbjct: 781 DPWHNN 798
>TC76410 similar to GP|4689382|gb|AAD27878.1| chlorophyll a/b binding
protein CP29 {Vigna radiata}, complete
Length = 1138
Score = 77.4 bits (189), Expect = 8e-16
Identities = 38/66 (57%), Positives = 48/66 (72%), Gaps = 1/66 (1%)
Frame = +2
Query: 21 LYPGGSF-DPLGLADDPEAFAELKVKELKNGRLAMFSMFGFFVQAIVTEKGPLENLADHI 79
LYPGG F DPLGLA DPE A L++ E+K+ RLAM + GF VQA T KGPL N A H+
Sbjct: 686 LYPGGKFFDPLGLASDPEKKATLQLAEIKHARLAMVAFLGFAVQAAATGKGPLNNWATHL 865
Query: 80 ADPVNS 85
+DP+++
Sbjct: 866 SDPLHT 883
>TC79544 similar to PIR|F96510|F96510 light-harvesting complex protein
[imported] - Arabidopsis thaliana, partial (82%)
Length = 1076
Score = 74.3 bits (181), Expect = 7e-15
Identities = 35/66 (53%), Positives = 50/66 (75%), Gaps = 1/66 (1%)
Frame = +1
Query: 18 VDPLYPGGSF-DPLGLADDPEAFAELKVKELKNGRLAMFSMFGFFVQAIVTEKGPLENLA 76
++P YPGG +PLGLA D ++ E K+KE+KNGRLAM ++ G FVQA VT GP++NL
Sbjct: 631 LEPGYPGGPLLNPLGLAKDIKSAREWKLKEIKNGRLAMVAILGIFVQASVTHVGPIDNLV 810
Query: 77 DHIADP 82
+H+++P
Sbjct: 811 EHLSNP 828
Score = 36.2 bits (82), Expect = 0.002
Identities = 20/54 (37%), Positives = 27/54 (49%), Gaps = 1/54 (1%)
Frame = +1
Query: 18 VDPLYPGG-SFDPLGLADDPEAFAELKVKELKNGRLAMFSMFGFFVQAIVTEKG 70
+D PG FDPLGL +DPE+ EL + R AM + G V ++ G
Sbjct: 280 LDGTLPGDFGFDPLGLGEDPESLKWYVQAELVHSRFAMLGVLGILVTDLLRVAG 441
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.319 0.141 0.426
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,043,288
Number of Sequences: 36976
Number of extensions: 16973
Number of successful extensions: 148
Number of sequences better than 10.0: 57
Number of HSP's better than 10.0 without gapping: 140
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 145
length of query: 98
length of database: 9,014,727
effective HSP length: 74
effective length of query: 24
effective length of database: 6,278,503
effective search space: 150684072
effective search space used: 150684072
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 50 (23.9 bits)
Medicago: description of AC139600.2


