

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC139525.8 - phase: 0
(676 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
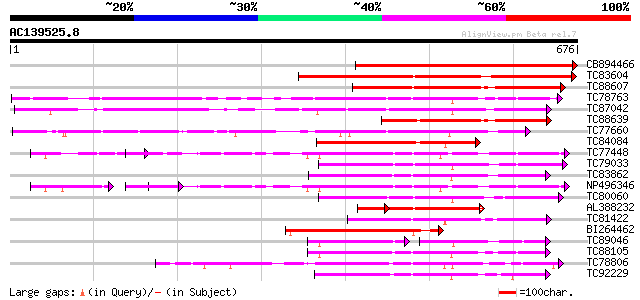
Score E
Sequences producing significant alignments: (bits) Value
CB894466 similar to PIR|B84782|B84 probable receptor-like protei... 526 e-149
TC83604 similar to PIR|T05606|T05606 protein kinase homolog F9D1... 327 7e-90
TC88607 similar to GP|8777331|dbj|BAA96921.1 receptor-like prote... 262 4e-70
TC78763 somatic embryogenesis receptor kinase 1 [Medicago trunca... 235 5e-62
TC87042 similar to GP|13605827|gb|AAK32899.1 AT5g48380/MJE7_1 {A... 231 7e-61
TC88639 similar to GP|22748334|gb|AAN05336.1 Putative leucine-ri... 200 1e-51
TC77660 similar to GP|22212524|dbj|BAC07504. receptor-like prote... 192 5e-49
TC84084 similar to GP|15810127|gb|AAL07207.1 putative receptor-k... 183 2e-46
TC77448 SYMRK; MtSYMRK [Medicago truncatula]; nod region linked ... 181 8e-46
TC79033 similar to GP|21554229|gb|AAM63304.1 somatic embryogenes... 173 2e-43
TC83862 similar to PIR|T05270|T05270 probable serine/threonine-s... 169 2e-42
NP496346 NP496346|AJ418371.1|CAD10810.1 nod region linked recept... 169 4e-42
TC80060 similar to PIR|D96574|D96574 hypothetical protein T3F20.... 167 1e-41
AL388232 weakly similar to GP|8777331|dbj receptor-like protein ... 137 2e-41
TC81422 similar to PIR|T02154|T02154 protein kinase homolog T1F1... 166 4e-41
BI264462 similar to GP|8777331|dbj receptor-like protein kinase ... 162 5e-40
TC89046 weakly similar to GP|21740816|emb|CAD41006. OSJNBa0042L1... 94 1e-39
TC88105 similar to GP|21593085|gb|AAM65034.1 Putative protein ki... 159 3e-39
TC78806 similar to GP|8978273|dbj|BAA98164.1 receptor protein ki... 158 6e-39
TC92229 similar to PIR|B86369|B86369 hypothetical protein AAC980... 157 1e-38
>CB894466 similar to PIR|B84782|B84 probable receptor-like protein kinase
[imported] - Arabidopsis thaliana, partial (36%)
Length = 871
Score = 526 bits (1354), Expect = e-149
Identities = 263/264 (99%), Positives = 263/264 (99%)
Frame = +3
Query: 413 KLKHPNIVKLRAYYYAKEEKLLVYDYLSNGSLHALLHGNRGPGRIPLDWTTRISLVLGAA 472
KLKHPNIVKL AYYYAKEEKLLVYDYLSNGSLHALLHGNRGPGRIPLDWTTRISLVLGAA
Sbjct: 3 KLKHPNIVKLLAYYYAKEEKLLVYDYLSNGSLHALLHGNRGPGRIPLDWTTRISLVLGAA 182
Query: 473 RGLARIHTEYSAAKVPHGNVKSSNVLLDKNGVACISDFGLSLLLNPVHATARLGGYRAPE 532
RGLARIHTEYSAAKVPHGNVKSSNVLLDKNGVACISDFGLSLLLNPVHATARLGGYRAPE
Sbjct: 183 RGLARIHTEYSAAKVPHGNVKSSNVLLDKNGVACISDFGLSLLLNPVHATARLGGYRAPE 362
Query: 533 QTEQKRLSQQADVYSFGVLLLEVLTGKAPSLQYPSPANRPRKVEEEETVVDLPKWVRSVV 592
QTEQKRLSQQADVYSFGVLLLEVLTGKAPSLQYPSPANRPRKVEEEETVVDLPKWVRSVV
Sbjct: 363 QTEQKRLSQQADVYSFGVLLLEVLTGKAPSLQYPSPANRPRKVEEEETVVDLPKWVRSVV 542
Query: 593 REEWTGEVFDQELLRYKNIEEELVSMLHVGLACVVQQPEKRPTMVDVVKMIEDIRVEQSP 652
REEWTGEVFDQELLRYKNIEEELVSMLHVGLACVVQQPEKRPTMVDVVKMIEDIRVEQSP
Sbjct: 543 REEWTGEVFDQELLRYKNIEEELVSMLHVGLACVVQQPEKRPTMVDVVKMIEDIRVEQSP 722
Query: 653 LCEDYDESRNSLSPSIPTTEDGLA 676
LCEDYDESRNSLSPSIPTTEDGLA
Sbjct: 723 LCEDYDESRNSLSPSIPTTEDGLA 794
>TC83604 similar to PIR|T05606|T05606 protein kinase homolog F9D16.210 -
Arabidopsis thaliana, partial (48%)
Length = 1142
Score = 327 bits (839), Expect = 7e-90
Identities = 164/333 (49%), Positives = 232/333 (69%), Gaps = 2/333 (0%)
Frame = +1
Query: 345 SKLVFFDRRN-GFELEDLLRASAEMLGKGSLGTVYRAVLDDGSTVAVKRLKDANPCARHE 403
+KL FF+ N F+LEDLLRASAE+LGKG+ GT Y+A+L+D + V VKRLK+ + +
Sbjct: 79 NKLTFFEGCNYAFDLEDLLRASAEVLGKGTFGTAYKAILEDATAVVVKRLKEV-AFGKKD 255
Query: 404 FEQYMDVIGKLKHPNIVKLRAYYYAKEEKLLVYDYLSNGSLHALLHGNRGPGRIPLDWTT 463
FEQYM+++G LKH N+V+L+AYYY+K+EKL+VYDY S GS+ +LLHG RG ++PLDW T
Sbjct: 256 FEQYMEIVGSLKHENVVELKAYYYSKDEKLMVYDYYSRGSVSSLLHGKRGEDKVPLDWDT 435
Query: 464 RISLVLGAARGLARIHTEYSAAKVPHGNVKSSNVLLDKNGVACISDFGLSLLLNPVH-AT 522
R+ + LGAARG+A+IH E + K+ HGN+KSSN+ L+ C+SD GL+ + +
Sbjct: 436 RLRIALGAARGIAQIHVE-NGGKLVHGNIKSSNIFLNTKQYGCVSDLGLATISTSLALPI 612
Query: 523 ARLGGYRAPEQTEQKRLSQQADVYSFGVLLLEVLTGKAPSLQYPSPANRPRKVEEEETVV 582
+R GYRAPE T+ ++ +Q +DVYSFGV+LLE+LTGK+P + ++
Sbjct: 613 SRAAGYRAPEVTDTRKAAQPSDVYSFGVVLLELLTGKSPI-----------HTTGGDEII 759
Query: 583 DLPKWVRSVVREEWTGEVFDQELLRYKNIEEELVSMLHVGLACVVQQPEKRPTMVDVVKM 642
L +WV SVVREEWT EVFD EL+RY NIEEE+V ML + ++CVV+ P++RP M +VVKM
Sbjct: 760 HLVRWVHSVVREEWTAEVFDLELMRYPNIEEEMVEMLQIAMSCVVRMPDQRPKMSEVVKM 939
Query: 643 IEDIRVEQSPLCEDYDESRNSLSPSIPTTEDGL 675
IE++R + + E++ + S T D +
Sbjct: 940 IENVRQIDNTQTQPSSENQGGVKLSSQTDYDNI 1038
>TC88607 similar to GP|8777331|dbj|BAA96921.1 receptor-like protein kinase
{Arabidopsis thaliana}, partial (35%)
Length = 899
Score = 262 bits (669), Expect = 4e-70
Identities = 135/255 (52%), Positives = 177/255 (68%), Gaps = 1/255 (0%)
Frame = +3
Query: 409 DVIGKL-KHPNIVKLRAYYYAKEEKLLVYDYLSNGSLHALLHGNRGPGRIPLDWTTRISL 467
+++G + HPN+V LRAYYY+K+EKLLV DY NG+L LLHG R GR LDW TR+ +
Sbjct: 3 EIVGSIGNHPNVVPLRAYYYSKDEKLLVCDYFPNGNLSILLHGTRTGGRTTLDWNTRVKI 182
Query: 468 VLGAARGLARIHTEYSAAKVPHGNVKSSNVLLDKNGVACISDFGLSLLLNPVHATARLGG 527
LG ARG+A +H + HGNVKSSNVLL+++ CISDFGL+ L+N +R G
Sbjct: 183 SLGIARGIAHLHL-VGGPRFTHGNVKSSNVLLNQDNDGCISDFGLTPLMNIPATPSRTMG 359
Query: 528 YRAPEQTEQKRLSQQADVYSFGVLLLEVLTGKAPSLQYPSPANRPRKVEEEETVVDLPKW 587
YRAPE E ++ + ++DVYSFGVLLLE+LTGKAP SP + +VDLP+W
Sbjct: 360 YRAPEVIETRKHTHKSDVYSFGVLLLEMLTGKAPQ---QSPVR--------DDMVDLPRW 506
Query: 588 VRSVVREEWTGEVFDQELLRYKNIEEELVSMLHVGLACVVQQPEKRPTMVDVVKMIEDIR 647
VRSVVREEWT EVFD EL+RY+NIEEE+V ML +G+ CV + P+ RP M +VV+MIE+IR
Sbjct: 507 VRSVVREEWTAEVFDVELMRYQNIEEEMVQMLQIGMTCVAKVPDMRPNMEEVVRMIEEIR 686
Query: 648 VEQSPLCEDYDESRN 662
S D++++
Sbjct: 687 QSDSDNRPSSDDNKS 731
>TC78763 somatic embryogenesis receptor kinase 1 [Medicago truncatula]
Length = 2737
Score = 235 bits (599), Expect = 5e-62
Identities = 197/664 (29%), Positives = 308/664 (45%), Gaps = 7/664 (1%)
Frame = +3
Query: 3 YVIMFFFFLFLSIYIVPCLTHNDTQALTLFRQQTDTHGQLLTNWTGPEACSASWHGVTCT 62
++ FF L +++V D AL R +L +W +W VTC
Sbjct: 453 FICAFFLLLLHPLWLVSANMEGD--ALHNLRTNLQDPNNVLQSWDPTLVNPCTWFHVTCN 626
Query: 63 PNNRVTTLVLPSLNLRGPIDALSSLTHLRLLDLHNNRLNGTVSASLLSNCTNLKLLYLAG 122
+N V + DL N L+GT+ L NL+ L L
Sbjct: 627 NDNSVIRV-----------------------DLGNAALSGTLVPQL-GQLKNLQYLELYS 734
Query: 123 NDFSGQIPPEISSLNNLLRLDLSDNNLAGDIPNEISRLTNLLTLRLQNNALSGNIPDLSS 182
N+ +G IP ++ +L NL+ LDL N G IP+ + +L+ L LRL NN+L G IP +
Sbjct: 735 NNITGPIPSDLGNLTNLVSLDLYLNRFNGPIPDSLGKLSKLRFLRLNNNSLMGPIPMSLT 914
Query: 183 IMPNLTELNMTNNEFYGKVP-NTMLNKFGDESFSGNEGLCGSKPFQVCSLTENSPPSSEP 241
+ L L+++NN+ G VP N + F SF+ N LCG C SPP S P
Sbjct: 915 NISALQVLDLSNNQLSGVVPDNGSFSLFTPISFANNLNLCGPVTGHPCP---GSPPFSPP 1085
Query: 242 VQTVPSNPSSFPATSVIARPRSQHHKGLSPGVIVAIVVAICVALLVVTSFVVAHCCARGR 301
VP P I+ P S G G + A A L+ + +A R R
Sbjct: 1086 PPFVPPPP--------ISAPGSGGATGAIAGGVAA------GAALLFAAPAIAFAWWRRR 1223
Query: 302 GVNSNSLMGSEAGKRKSYGSEKKVYNSNGGGGDSSDGTSGTDMSKLVFFDRRNGFELEDL 361
K + + + ++ + +L F R D
Sbjct: 1224 -------------KPQEFFFDVP-----------AEEDPEVHLGQLKRFSLRELQVATDT 1331
Query: 362 LRASAEMLGKGSLGTVYRAVLDDGSTVAVKRLKDAN-PCARHEFEQYMDVIGKLKHPNIV 420
++ +LG+G G VY+ L DGS VAVKRLK+ P +F+ +++I H N++
Sbjct: 1332 F-SNKNILGRGGFGKVYKGRLADGSLVAVKRLKEERTPGGELQFQTEVEMISMAVHRNLL 1508
Query: 421 KLRAYYYAKEEKLLVYDYLSNGSLHALLHGNRGPGRIPLDWTTRISLVLGAARGLARIHT 480
+LR + E+LLVY Y++NGS+ + L R P + PLDW TR + LG+ARGL+ +H
Sbjct: 1509 RLRGFCMTPTERLLVYPYMANGSVASCLR-ERPPHQEPLDWPTRKRIALGSARGLSYLH- 1682
Query: 481 EYSAAKVPHGNVKSSNVLLDKNGVACISDFGLSLLLN--PVHATARLG---GYRAPEQTE 535
++ K+ H +VK++N+LLD+ A + DFGL+ L++ H T + G+ APE
Sbjct: 1683 DHCDPKIIHRDVKAANILLDEEFEAVVGDFGLAKLMDYKDTHVTTAVRGTIGHIAPEYLS 1862
Query: 536 QKRLSQQADVYSFGVLLLEVLTGKAPSLQYPSPANRPRKVEEEETVVDLPKWVRSVVREE 595
+ S++ DV+ +G++LLE++TG+ + R +++ V L WV+ +++E+
Sbjct: 1863 TGKSSEKTDVFGYGIMLLELITGQ-------RAFDLARLANDDD--VMLLDWVKGLLKEK 2015
Query: 596 WTGEVFDQELLRYKNIEEELVSMLHVGLACVVQQPEKRPTMVDVVKMIEDIRVEQSPLCE 655
+ D + L+ IE E+ ++ V L C P RP M DVV+M+E L E
Sbjct: 2016 KLEMLVDPD-LKTNYIEAEVEQLIQVALLCTQGSPMDRPKMSDVVRMLEG-----DGLAE 2177
Query: 656 DYDE 659
+DE
Sbjct: 2178 RWDE 2189
>TC87042 similar to GP|13605827|gb|AAK32899.1 AT5g48380/MJE7_1 {Arabidopsis
thaliana}, partial (51%)
Length = 2206
Score = 231 bits (589), Expect = 7e-61
Identities = 193/661 (29%), Positives = 300/661 (45%), Gaps = 20/661 (3%)
Frame = +2
Query: 6 MFFFFLFLSIYIVPCLTHNDTQALTLFRQQT-DTHGQLLTNWT---GPEACSASWHGVTC 61
+F FL L + + T D L + D + L ++W E ++GV C
Sbjct: 185 VFVSFLLLISFGITYGTETDIFCLKSIKNSIQDPNNYLTSSWNFNNKTEGFICRFNGVEC 364
Query: 62 --TPNNRVTTLVLPSLNLRGPIDALSSLTHLRLLDLHNNRLNGTVSASLLSNCTNLKLLY 119
N+V L L ++ L+G G V NC+++ L
Sbjct: 365 WHPDENKVLNLKLSNMGLKGQFP------------------RGIV------NCSSMTGLD 472
Query: 120 LAGNDFSGQIPPEISSLNNLLR-LDLSDNNLAGDIPNEISRLTNLLTLRLQNNALSGNIP 178
L+ ND SG IP +IS+L + LDLS N +G+IP ++ T L L+L N L+G IP
Sbjct: 473 LSVNDLSGTIPGDISTLLKFVTSLDLSSNEFSGEIPVSLANCTYLNVLKLSQNQLTGQIP 652
Query: 179 DLSSIMPNLTELNMTNNEFYGKVPNTMLNKFGDESFSGNEGLCGSKPFQVCSLTENSPPS 238
L + + +++NN G+VPN D +++ N+GLCG VC T +S +
Sbjct: 653 LLLGTLDRIKTFDVSNNLLTGQVPNFTAGGKVDVNYANNQGLCGQPSLGVCKATASSKSN 832
Query: 239 SEPVQTVPSNPSSFPATSVIARPRSQHHKGLSPGVIVAIVVAICVALLVVTSFVVAHCCA 298
T+VIA G V V + L V F V
Sbjct: 833 ----------------TAVIA------------GAAVGAVTLAALGLGVFMFFFV----- 913
Query: 299 RGRGVNSNSLMGSEAGKRKSYGSEKKVYNSNGGGGDSSDGTSGTDMSKLVFFDRRNGFEL 358
R+S +K+ S GT G +S +F + +L
Sbjct: 914 -----------------RRSAYRKKEEDPEGNKWARSLKGTKGIKVS--LFEKSISKMKL 1036
Query: 359 EDLLRAS-----AEMLGKGSLGTVYRAVLDDGSTVAVKRLKDANPCARHEFEQYMDVIGK 413
DL++A+ ++G G GTVY+A L+DG+ VKRL+++ + EF M +G
Sbjct: 1037SDLMKATNNFSNINIIGTGRTGTVYKATLEDGTAFMVKRLQESQHSEK-EFMSEMATLGT 1213
Query: 414 LKHPNIVKLRAYYYAKEEKLLVYDYLSNGSLHALLHGNRGPGRIPLDWTTRISLVLGAAR 473
+KH N+V L + AK+E+LLV+ + NG LH LH G LDW +R+ + +GAA+
Sbjct: 1214VKHRNLVPLLGFCVAKKERLLVFKNMPNGMLHDQLH--PAAGECTLDWPSRLKIAIGAAK 1387
Query: 474 GLARIHTEYSAAKVPHGNVKSSNVLLDKNGVACISDFGLSLLLNPV--HATARLG----- 526
G A +H + ++ H N+ S +LLD + ISDFGL+ L+NP+ H + +
Sbjct: 1388GFAWLHHSCN-PRIIHRNISSKCILLDADFEPKISDFGLARLMNPLDTHLSTFVNGEFGD 1564
Query: 527 -GYRAPEQTEQKRLSQQADVYSFGVLLLEVLTGKAPSLQYPSPANRPRKVEEEETVVDLP 585
GY APE T+ + + DV+SFG +LLE++TG+ P+ +P E +L
Sbjct: 1565FGYVAPEYTKTLVATPKGDVFSFGTVLLELVTGERPANVAKAP---------ETFKGNLV 1717
Query: 586 KWVRSVVREEWTGEVFDQELLRYKNIEEELVSMLHVGLACVVQQPEKRPTMVDVVKMIED 645
+W+ + + D+ LL K + EL L V CV + P++RPTM +V + +
Sbjct: 1718EWITELSSNSKLHDAIDESLLN-KGDDNELFQFLKVACNCVTEVPKERPTMFEVYQFLRA 1894
Query: 646 I 646
I
Sbjct: 1895I 1897
>TC88639 similar to GP|22748334|gb|AAN05336.1 Putative leucine-rich repeat
transmembrane protein kinase {Oryza sativa (japonica
cultivar-group)}, partial (28%)
Length = 869
Score = 200 bits (509), Expect = 1e-51
Identities = 103/203 (50%), Positives = 141/203 (68%)
Frame = +1
Query: 444 LHALLHGNRGPGRIPLDWTTRISLVLGAARGLARIHTEYSAAKVPHGNVKSSNVLLDKNG 503
L ALLH N G GR PL+W TR ++ LGAA+G+A +H++ + HGN+KSSN+LL K+
Sbjct: 1 LSALLHANNGAGRTPLNWETRSTIALGAAQGIAYLHSQSPTSS--HGNIKSSNILLTKSF 174
Query: 504 VACISDFGLSLLLNPVHATARLGGYRAPEQTEQKRLSQQADVYSFGVLLLEVLTGKAPSL 563
+SDFGL+ L P R+ GYRAPE T+ +++SQ+ADVYSFG++LLE+LTGKAP+
Sbjct: 175 EPRVSDFGLAYLALPTATPNRVSGYRAPEVTDARKVSQKADVYSFGIMLLELLTGKAPT- 351
Query: 564 QYPSPANRPRKVEEEETVVDLPKWVRSVVREEWTGEVFDQELLRYKNIEEELVSMLHVGL 623
S N E VDLP+WV+S+V++EW EVFD ELLRY+++EEE+V++L + L
Sbjct: 352 --HSSLN--------EEGVDLPRWVQSIVQDEWNTEVFDMELLRYQSVEEEMVNLLQLAL 501
Query: 624 ACVVQQPEKRPTMVDVVKMIEDI 646
C Q P+KRP+M V IE I
Sbjct: 502 ECTTQYPDKRPSMDVVASKIEKI 570
>TC77660 similar to GP|22212524|dbj|BAC07504. receptor-like protein kinase
{Nicotiana tabacum}, partial (69%)
Length = 1886
Score = 192 bits (487), Expect = 5e-49
Identities = 184/655 (28%), Positives = 292/655 (44%), Gaps = 37/655 (5%)
Frame = +2
Query: 4 VIMFFFFLFLSIYIVPCLTHNDTQALTLFRQQTDTHGQLLTNWTGPEACSASWHGVTCT- 62
+++ F L SI+ L +DT+AL+ + + TN C+ GV C
Sbjct: 104 LLLSFLSLLFSIHAKLTLHPSDTKALSTLQNNLGLNLDTTTN----NLCNKE--GVFCER 265
Query: 63 --PNN-----RVTTLVLPSLNLRGPID-ALSSLTHLRLLDLHNNRLNGTVSASLLSNCTN 114
NN RVT LV S L G + + LT L+ + L +N+L + S++ +C
Sbjct: 266 RLTNNESYALRVTKLVFKSRKLSGILSPTIGKLTELKEISLSDNKLVDQIPTSIV-DCRK 442
Query: 115 LKLLYLAGNDFSGQIPPEISSLNNLLRLDLSDNNLAGDIPNEISRLTNLLTLRLQNNALS 174
L+ L LA N FSG++P E SSL L LD+S N L+G++ N + NL TL + +N +
Sbjct: 443 LEFLNLANNLFSGEVPSEFSSLIRLRFLDISGNKLSGNL-NFLRYFPNLETLSVADNHFT 619
Query: 175 GNIPDLSSIMPNLTELNMTNNEFYGKVPNTMLNKFGDESFSGNEGLCGSKPFQVCSLTEN 234
G +P NL N + N F VP LN + G E + P + N
Sbjct: 620 GRVPVSVRSFRNLRHFNFSGNRFLEGVP---LN----QKLLGYEDTDNTAPKRYILAETN 778
Query: 235 SPPSSEPVQTVPSNPSSFPATSVIARPRSQHHK------GLSPGVIVAIVVAICVALLVV 288
+ + P ++ +P + PA + A P +H K G G + I +
Sbjct: 779 NSSQTRPHRS--HSPGAAPAPAPAA-PLHKHKKSRRKLAGWILGFVAGAFAGILSGFVFS 949
Query: 289 TSFVVAHCCARGRGVNSNSLMGSEAGKRKSYGSEKKVYNSNGGGGDSSDGTSGTDMSKLV 348
F +A +G+G S + S K+ L
Sbjct: 950 LLFKLALILIKGKGKGSGPAIYSSLIKKAE---------------------------DLA 1048
Query: 349 FFDRRNGFELEDLLRASAEMLGKGSLGTVYRAVL--DDGSTVAVKRL----KDANPCARH 402
F ++ +G AS E +G+G VY+A L +G +A+K++ KDA A
Sbjct: 1049FLEKEDGL-------ASLEKIGQGGCVEVYKAELPGSNGKMIAIKKIIQPPKDAAELAEE 1207
Query: 403 E----------FEQYMDVIGKLKHPNIVKLRAYYYAKEEKLLVYDYLSNGSLHALLHGNR 452
+ + +D +G+++H N++ L A+ + LVY+++ NGSL +LH
Sbjct: 1208DSKLLHKKMRQIKSEIDTVGQIRHRNLLPLLAHISRPDCHYLVYEFMKNGSLQDMLH-KV 1384
Query: 453 GPGRIPLDWTTRISLVLGAARGLARIHTEYSAAKVPHGNVKSSNVLLDKNGVACISDFGL 512
G LDW R + LG A GL +HT +S ++ H ++K +NVLLD A I+DFGL
Sbjct: 1385ERGEAELDWLARHKIALGIAAGLEYLHTSHS-PRIIHRDLKPANVLLDDEMEARIADFGL 1561
Query: 513 SLLL--NPVHAT----ARLGGYRAPEQTEQKRLSQQADVYSFGVLLLEVLTGKAPSLQYP 566
+ + H T A GY APE + + + + D+YSFGV+L ++ GK PS +
Sbjct: 1562AKAMPDAQTHITTSNVAGTVGYIAPEYHQILKFNDKCDIYSFGVMLGVLVIGKLPSDDFF 1741
Query: 567 SPANRPRKVEEEETVVDLPKWVRSVVREEWTGEVFDQELLRYKNIEEELVSMLHV 621
+ + + L KW+R+V+ E E D LL EE+++ +L +
Sbjct: 1742TNTDE----------MSLVKWMRNVMTSENPKEAIDARLLS-NGFEEQMLLVLXI 1873
>TC84084 similar to GP|15810127|gb|AAL07207.1 putative receptor-kinase
isolog {Arabidopsis thaliana}, partial (29%)
Length = 595
Score = 183 bits (465), Expect = 2e-46
Identities = 101/200 (50%), Positives = 135/200 (67%), Gaps = 5/200 (2%)
Frame = +1
Query: 367 EMLGKGSLGTVYRAVLDDGSTVAVKRLKDANPCARHEFEQYMDVIGKLKHPNIVKLRAYY 426
E LG+G +G+ Y+AV++ G V VKRLKDA EF ++D++GKL+HPN+V LRAY+
Sbjct: 1 ETLGRGIMGSTYKAVMESGFIVTVKRLKDARYPGLEEFRAHIDLLGKLRHPNLVPLRAYF 180
Query: 427 YAKEEKLLVYDYLSNGSLHALLHGNR-GPGRIPLDWTTRISLVLGAARGLARIHTEYSAA 485
AKEE+LLVYDY NGSL +L+HG + G PL WT+ + + A GL IH A
Sbjct: 181 QAKEERLLVYDYFPNGSLFSLVHGTKTSSGGKPLHWTSCLKIAEDLATGLLYIHQNPGMA 360
Query: 486 KVPHGNVKSSNVLLDKNGVACISDFGLSLLLNP---VHATARLGGYRAPE-QTEQKRLSQ 541
HGN+KSSNVLL + +C++D+GL++ LNP +A YRAPE ++ Q+ +Q
Sbjct: 361 ---HGNLKSSNVLLGADFESCLTDYGLTVFLNPDTMEEPSATSFFYRAPECRSFQRPQTQ 531
Query: 542 QADVYSFGVLLLEVLTGKAP 561
ADVYSFGVLLLE+LTGK P
Sbjct: 532 PADVYSFGVLLLELLTGKTP 591
>TC77448 SYMRK; MtSYMRK [Medicago truncatula]; nod region linked receptor
kinase [Medicago truncatula]
Length = 3568
Score = 181 bits (459), Expect = 8e-46
Identities = 161/546 (29%), Positives = 258/546 (46%), Gaps = 17/546 (3%)
Frame = +1
Query: 139 LLRLDLSDNNLAGDIPNEISRLTNLLTLRLQNNALSGNIPDL--SSIMPNLTELNMTNNE 196
+ +LDLS NNL G IP+ ++++TNL L L +N P SS+ L L+++ N+
Sbjct: 1597 ITKLDLSSNNLKGAIPSIVTKMTNLQILNLSHNQFDMLFPSFPPSSL---LISLDLSYND 1767
Query: 197 FYGKVPNTMLNKFGDESFSGNEGLCGSKPFQVCSLTENSPPSSEPVQTVPSNPSSFPATS 256
G +P +++ S P + SL PS T N S +
Sbjct: 1768 LSGWLPESII----------------SLP-HLKSLYFGCNPSMSDEDTTKLNSSLI--NT 1890
Query: 257 VIARPRSQHHKGLSPGVIVAIVVAICVALLVVTSFVVAHCCARGRGVNSNSLMGSEAGKR 316
R +++ K VI AI + L V ++ C R + + G
Sbjct: 1891 DYGRCKAKKPKFGQVFVIGAITSGSLLITLAVG--ILFFCRYRHKSITLE-------GFG 2043
Query: 317 KSYGSEKKVYNSNGGGGDSSDGTSGTDMSKLVFFDRR---NGFELEDLLRASAE---MLG 370
K+Y + S D FF + F LE + +A+ + ++G
Sbjct: 2044 KTYPMATNIIFSLPSKDD--------------FFIKSVSVKPFTLEYIEQATEQYKTLIG 2181
Query: 371 KGSLGTVYRAVLDDGSTVAVKRLKDANPCARHEFEQYMDVIGKLKHPNIVKLRAYYYAKE 430
+G G+VYR LDDG VAVK + EF+ ++++ ++H N+V L Y +
Sbjct: 2182 EGGFGSVYRGTLDDGQEVAVKVRSSTSTQGTREFDNELNLLSAIQHENLVPLLGYCNEYD 2361
Query: 431 EKLLVYDYLSNGSLHALLHGNRGPGRIPLDWTTRISLVLGAARGLARIHTEYSAAKVPHG 490
+++LVY ++SNGSL L+G +I LDW TR+S+ LGAARGLA +HT + V H
Sbjct: 2362 QQILVYPFMSNGSLLDRLYGEASKRKI-LDWPTRLSIALGAARGLAYLHT-FPGRSVIHR 2535
Query: 491 NVKSSNVLLDKNGVACISDFGL---------SLLLNPVHATARLGGYRAPEQTEQKRLSQ 541
+VKSSN+LLD++ A ++DFG S + V TA GY PE + ++LS+
Sbjct: 2536 DVKSSNILLDQSMCAKVADFGFSKYAPQEGDSYVSLEVRGTA---GYLDPEYYKTQQLSE 2706
Query: 542 QADVYSFGVLLLEVLTGKAPSLQYPSPANRPRKVEEEETVVDLPKWVRSVVREEWTGEVF 601
++DV+SFGV+LLE+++G+ P ++ L +W + +R E+
Sbjct: 2707 KSDVFSFGVVLLEIVSGR-----------EPLNIKRPRIEWSLVEWAKPYIRASKVDEIV 2853
Query: 602 DQELLRYKNIEEELVSMLHVGLACVVQQPEKRPTMVDVVKMIEDIRVEQSPLCEDYDESR 661
D ++ E L ++ V L C+ RP MVD+V+ +ED + ++ E Y +S
Sbjct: 2854 DPG-IKGGYHAEALWRVVEVALQCLEPYSTYRPCMVDIVRELEDALIIENNASE-YMKSI 3027
Query: 662 NSLSPS 667
+SL S
Sbjct: 3028 DSLGGS 3045
Score = 52.8 bits (125), Expect = 4e-07
Identities = 40/145 (27%), Positives = 66/145 (44%), Gaps = 3/145 (2%)
Frame = +1
Query: 25 DTQALTLFRQQTDTHGQ---LLTNWTGPEACSASWHGVTCTPNNRVTTLVLPSLNLRGPI 81
D + + R++ H Q L +W+G W G+TC
Sbjct: 1453 DLEVIQKMREELLLHNQENEALESWSGDPCMIFPWKGITC-------------------- 1572
Query: 82 DALSSLTHLRLLDLHNNRLNGTVSASLLSNCTNLKLLYLAGNDFSGQIPPEISSLNNLLR 141
D + + + LDL +N L G + S+++ TNL++L L+ N F + P + L+
Sbjct: 1573 DDSTGSSIITKLDLSSNNLKGAI-PSIVTKMTNLQILNLSHNQFD-MLFPSFPPSSLLIS 1746
Query: 142 LDLSDNNLAGDIPNEISRLTNLLTL 166
LDLS N+L+G +P I L +L +L
Sbjct: 1747 LDLSYNDLSGWLPESIISLPHLKSL 1821
>TC79033 similar to GP|21554229|gb|AAM63304.1 somatic embryogenesis
receptor-like kinase putative {Arabidopsis thaliana},
partial (77%)
Length = 1762
Score = 173 bits (439), Expect = 2e-43
Identities = 108/302 (35%), Positives = 169/302 (55%), Gaps = 5/302 (1%)
Frame = +1
Query: 369 LGKGSLGTVYRAVLDDGSTVAVKRLKDANPCARHEFEQYMDVIGKLKHPNIVKLRAYYYA 428
LG+G G+VY L DGS +AVKRLK + A EF ++++ +++H N++ LR Y
Sbjct: 481 LGEGGFGSVYWGQLWDGSQIAVKRLKVWSNKADMEFAVEVEILARVRHKNLLSLRGYCAE 660
Query: 429 KEEKLLVYDYLSNGSLHALLHGNRGPGRIPLDWTTRISLVLGAARGLARIHTEYSAAKVP 488
+E+L+VYDY+ N SL + LHG + LDW R+++ +G+A G+ +H + + +
Sbjct: 661 GQERLIVYDYMPNLSLLSHLHGQHSTESL-LDWNRRMNIAIGSAEGIVYLHVQ-ATPHII 834
Query: 489 HGNVKSSNVLLDKNGVACISDFGLSLLL--NPVHATARLG---GYRAPEQTEQKRLSQQA 543
H +VK+SNVLLD + A ++DFG + L+ H T R+ GY APE + ++
Sbjct: 835 HRDVKASNVLLDSDFQARVADFGFAKLIPDGATHVTTRVKGTLGYLAPEYAMLGKANESC 1014
Query: 544 DVYSFGVLLLEVLTGKAPSLQYPSPANRPRKVEEEETVVDLPKWVRSVVREEWTGEVFDQ 603
DVYSFG+LLLE+ +GK P + S R + W + E+ E+ D
Sbjct: 1015DVYSFGILLLELASGKKPLEKLSSSVKRA-----------INDWALPLACEKKFSELADP 1161
Query: 604 ELLRYKNIEEELVSMLHVGLACVVQQPEKRPTMVDVVKMIEDIRVEQSPLCEDYDESRNS 663
L +EEEL ++ V L C QPEKRPTMV+VV++++ E+ E+ + +N
Sbjct: 1162R-LNGDYVEEELKRVILVALICAQNQPEKRPTMVEVVELLKGESKEKVLQLENNELFKNP 1338
Query: 664 LS 665
L+
Sbjct: 1339LA 1344
>TC83862 similar to PIR|T05270|T05270 probable serine/threonine-specific
protein kinase (EC 2.7.1.-) T4L20.80 - Arabidopsis
thaliana, partial (60%)
Length = 948
Score = 169 bits (429), Expect = 2e-42
Identities = 112/295 (37%), Positives = 162/295 (53%), Gaps = 7/295 (2%)
Frame = +1
Query: 357 ELEDLLRASAE--MLGKGSLGTVYRAVLDDGSTVAVKRLKDANPCARHEFEQYMDVIGKL 414
ELE+ AE ++G+ G VYR +L DGS VAVK L + A EF+ ++ IGK+
Sbjct: 25 ELENATDGFAEGSVIGERGYGIVYRGILQDGSIVAVKNLLNNKGQAEKEFKVEVEAIGKV 204
Query: 415 KHPNIVKLRAYYYAKEEKLLVYDYLSNGSLHALLHGNRGPGRIPLDWTTRISLVLGAARG 474
+H N+V L Y +++LVY+Y+ NG+L LHG+ GP PL W R+ + +G A+G
Sbjct: 205 RHKNLVGLVGYCAEGAKRMLVYEYVDNGNLEQWLHGDVGPVS-PLTWDIRMKIAVGTAKG 381
Query: 475 LARIHTEYSAAKVPHGNVKSSNVLLDKNGVACISDFGLSLLL--NPVHATARL---GGYR 529
LA +H E KV H +VKSSN+LLDK A +SDFGL+ LL + T R+ GY
Sbjct: 382 LAYLH-EGLEPKVVHRDVKSSNILLDKKWHAKVSDFGLAKLLGSGKSYVTTRVMGTFGYV 558
Query: 530 APEQTEQKRLSQQADVYSFGVLLLEVLTGKAPSLQYPSPANRPRKVEEEETVVDLPKWVR 589
+PE L++ +DVYSFG+LL+E++TG++P +PA ++L W +
Sbjct: 559 SPEYASTGMLNEGSDVYSFGILLMELVTGRSPIDYSRAPAE-----------MNLVDWFK 705
Query: 590 SVVREEWTGEVFDQELLRYKNIEEELVSMLHVGLACVVQQPEKRPTMVDVVKMIE 644
+V GE L+ + L L V L C+ KRP M +V M+E
Sbjct: 706 GMVASR-RGEELVDPLIEIQPSPRSLKRALLVCLRCIDLDANKRPKMGQIVHMLE 867
>NP496346 NP496346|AJ418371.1|CAD10810.1 nod region linked receptor kinase
[Medicago truncatula]
Length = 2706
Score = 169 bits (427), Expect = 4e-42
Identities = 151/517 (29%), Positives = 242/517 (46%), Gaps = 15/517 (2%)
Frame = +1
Query: 166 LRLQNNALSGNIPDLSSIMPNLTELNMTNNEFYGKVPNTMLNKFGDESFSGNEGLCGSKP 225
L L +N L G IP + + M NL L+++ N+ G +P +++ S P
Sbjct: 1231 LDLSSNNLKGAIPSIVTKMTNLQILDLSYNDLSGWLPESII----------------SLP 1362
Query: 226 FQVCSLTENSPPSSEPVQTVPSNPSSFPATSVIARPRSQHHKGLSPGVIVAIVVAICVAL 285
+ SL PS T N S + R +++ K VI AI +
Sbjct: 1363 -HLKSLYFGCNPSMSDEDTTKLNSSLI--NTDYGRCKAKKPKFGQVFVIGAITSGSLLIT 1533
Query: 286 LVVTSFVVAHCCARGRGVNSNSLMGSEAGKRKSYGSEKKVYNSNGGGGDSSDGTSGTDMS 345
L V ++ C R + + G K+Y + S D
Sbjct: 1534 LAVG--ILFFCRYRHKSITLE-------GFGKTYPMATNIIFSLPSKDD----------- 1653
Query: 346 KLVFFDRR---NGFELEDLLRASAE---MLGKGSLGTVYRAVLDDGSTVAVKRLKDANPC 399
FF + F LE + +A+ + ++G+G G+VYR LDDG VAVK +
Sbjct: 1654 ---FFIKSVSVKPFTLEYIEQATEQYKTLIGEGGFGSVYRGTLDDGQEVAVKVRSSTSTQ 1824
Query: 400 ARHEFEQYMDVIGKLKHPNIVKLRAYYYAKEEKLLVYDYLSNGSLHALLHGNRGPGRIPL 459
EF+ ++++ ++H N+V L Y ++++LVY ++SNGSL L+G +I L
Sbjct: 1825 GTREFDNELNLLSAIQHENLVPLLGYCNEYDQQILVYPFMSNGSLLDRLYGEASKRKI-L 2001
Query: 460 DWTTRISLVLGAARGLARIHTEYSAAKVPHGNVKSSNVLLDKNGVACISDFGL------- 512
DW TR+S+ LGAARGLA +HT + V H +VKSSN+LLD++ A ++DFG
Sbjct: 2002 DWPTRLSIALGAARGLAYLHT-FPGRSVIHRDVKSSNILLDQSMCAKVADFGFSKYAPQE 2178
Query: 513 --SLLLNPVHATARLGGYRAPEQTEQKRLSQQADVYSFGVLLLEVLTGKAPSLQYPSPAN 570
S + V TA GY PE + ++LS+++DV+SFGV+LLE+++G+
Sbjct: 2179 GDSYVSLEVRGTA---GYLDPEYYKTQQLSEKSDVFSFGVVLLEIVSGR----------- 2316
Query: 571 RPRKVEEEETVVDLPKWVRSVVREEWTGEVFDQELLRYKNIEEELVSMLHVGLACVVQQP 630
P ++ L +W + +R E+ D ++ E L ++ V L C+
Sbjct: 2317 EPLNIKRPRIEWSLVEWAKPYIRASKVDEIVDPG-IKGGYHAEALWRVVEVALQCLEPYS 2493
Query: 631 EKRPTMVDVVKMIEDIRVEQSPLCEDYDESRNSLSPS 667
RP MVD+V+ +ED + ++ E Y +S +SL S
Sbjct: 2494 TYRPCMVDIVRELEDALIIENNASE-YMKSIDSLGGS 2601
Score = 52.4 bits (124), Expect = 6e-07
Identities = 35/106 (33%), Positives = 58/106 (54%), Gaps = 7/106 (6%)
Frame = +1
Query: 25 DTQALTLFRQQTDTHGQ---LLTNWTGPEACSASWHGVTC---TPNNRVTTLVLPSLNLR 78
D + + R++ H Q L +W+G W G+TC T ++ +T L L S NL+
Sbjct: 1078 DLEVIQKMREELLLHNQENEALESWSGDPCMIFPWKGITCDDSTGSSIITKLDLSSNNLK 1257
Query: 79 GPIDAL-SSLTHLRLLDLHNNRLNGTVSASLLSNCTNLKLLYLAGN 123
G I ++ + +T+L++LDL N L+G + S++S +LK LY N
Sbjct: 1258 GAIPSIVTKMTNLQILDLSYNDLSGWLPESIIS-LPHLKSLYFGCN 1392
Score = 47.8 bits (112), Expect = 1e-05
Identities = 27/69 (39%), Positives = 38/69 (54%)
Frame = +1
Query: 139 LLRLDLSDNNLAGDIPNEISRLTNLLTLRLQNNALSGNIPDLSSIMPNLTELNMTNNEFY 198
+ +LDLS NNL G IP+ ++++TNL L L N LSG +P+ +P+L L N
Sbjct: 1222 ITKLDLSSNNLKGAIPSIVTKMTNLQILDLSYNDLSGWLPESIISLPHLKSLYFGCNPSM 1401
Query: 199 GKVPNTMLN 207
T LN
Sbjct: 1402 SDEDTTKLN 1428
>TC80060 similar to PIR|D96574|D96574 hypothetical protein T3F20.24
[imported] - Arabidopsis thaliana, partial (32%)
Length = 1547
Score = 167 bits (423), Expect = 1e-41
Identities = 111/297 (37%), Positives = 159/297 (53%), Gaps = 5/297 (1%)
Frame = +1
Query: 369 LGKGSLGTVYRAVLDDGSTVAVKRLKDANPCARHEFEQYMDVIGKLKHPNIVKLRAYYYA 428
+G+G G VY+ VL DG+ +AVK+L + EF + +I L+HPN+VKL
Sbjct: 211 IGEGGFGPVYKGVLSDGAVIAVKQLSSKSKQGNREFVNEIGMISALQHPNLVKLYGCCIE 390
Query: 429 KEEKLLVYDYLSNGSLHALLHGNRGPGRIPLDWTTRISLVLGAARGLARIHTEYSAAKVP 488
+ LLVY+Y+ N SL L G + R+ LDW TR+ + +G ARGLA +H E S K+
Sbjct: 391 GNQLLLVYEYMENNSLARALFG-KPEQRLNLDWRTRMKICVGIARGLAYLHEE-SRLKIV 564
Query: 489 HGNVKSSNVLLDKNGVACISDFGLSLL--LNPVHATARLG---GYRAPEQTEQKRLSQQA 543
H ++K++NVLLDKN A ISDFGL+ L H + R+ GY APE + L+ +A
Sbjct: 565 HRDIKATNVLLDKNLNAKISDFGLAKLDEEENTHISTRIAGTIGYMAPEYAMRGYLTDKA 744
Query: 544 DVYSFGVLLLEVLTGKAPSLQYPSPANRPRKVEEEETVVDLPKWVRSVVREEWTGEVFDQ 603
DVYSFGV+ LE+++G + + P +E V L W V++E+
Sbjct: 745 DVYSFGVVALEIVSGMSNTNYRP-----------KEEFVYLLDWA-YVLQEQGNLLELVD 888
Query: 604 ELLRYKNIEEELVSMLHVGLACVVQQPEKRPTMVDVVKMIEDIRVEQSPLCEDYDES 660
L K EE + ML + L C P RP M VV M+E Q+P+ + D +
Sbjct: 889 PTLGSKYSSEEAMRMLQLALLCTNPSPTLRPPMSSVVSMLEGNTPIQAPIIKRSDST 1059
>AL388232 weakly similar to GP|8777331|dbj receptor-like protein kinase
{Arabidopsis thaliana}, partial (22%)
Length = 488
Score = 137 bits (344), Expect(2) = 2e-41
Identities = 67/119 (56%), Positives = 88/119 (73%)
Frame = +1
Query: 448 LHGNRGPGRIPLDWTTRISLVLGAARGLARIHTEYSAAKVPHGNVKSSNVLLDKNGVACI 507
LHGN+G GR P DW +R+ + LGAA+G+A IHTE K HGN+KS+NVL+ + +CI
Sbjct: 112 LHGNKGAGRTPFDWNSRVKVALGAAKGIAFIHTE-GGQKFTHGNIKSTNVLITEEFDSCI 288
Query: 508 SDFGLSLLLNPVHATARLGGYRAPEQTEQKRLSQQADVYSFGVLLLEVLTGKAPSLQYP 566
SD GL L+N +R GYRAPE T+ K+++Q++DVYSFGVLLLE+LTGK P L+YP
Sbjct: 289 SDVGLPPLMNAPATMSRTNGYRAPEVTDSKKITQKSDVYSFGVLLLELLTGKVP-LRYP 462
Score = 51.2 bits (121), Expect(2) = 2e-41
Identities = 21/39 (53%), Positives = 29/39 (73%)
Frame = +3
Query: 415 KHPNIVKLRAYYYAKEEKLLVYDYLSNGSLHALLHGNRG 453
+HPN++ LRAYYY+K+EKL+VY Y+ GSL L +G
Sbjct: 12 RHPNVMPLRAYYYSKDEKLMVYSYMPEGSLFFLFAWKQG 128
>TC81422 similar to PIR|T02154|T02154 protein kinase homolog T1F15.2 -
Arabidopsis thaliana, partial (34%)
Length = 1130
Score = 166 bits (419), Expect = 4e-41
Identities = 109/265 (41%), Positives = 148/265 (55%), Gaps = 21/265 (7%)
Frame = +2
Query: 403 EFEQYMDVIGKLKHPNIVKLRAYYYAKEEKLLVYDYLSNGSLHALLHGNRGPGRIPLDWT 462
EF + IGK+KHPNIVKLRAYY+A +EKLL+ D++SNG+L L G G L W+
Sbjct: 20 EFATEVQAIGKVKHPNIVKLRAYYWAHDEKLLISDFVSNGNLANALRGRNGQPSPNLSWS 199
Query: 463 TRISLVLGAARGLARIHTEYSAAKVPHGNVKSSNVLLDKNGVACISDFGLSLLL-----N 517
R+ + G ARGLA +H E S K HG++K SN+LLD + ISDFGL+ L+ N
Sbjct: 200 IRLRIAKGTARGLAYLH-ECSPRKFVHGDLKPSNILLDTDFQPLISDFGLNRLISITGNN 376
Query: 518 P--------------VHATARLGGYRAPE-QTEQKRLSQQADVYSFGVLLLEVLTGKAPS 562
P T R Y+APE + R +Q+ DVYSFGV+LLE+LTGK+P
Sbjct: 377 PSTGGFMGGALPYMKSSQTERTNNYKAPEAKVPGCRPTQKWDVYSFGVVLLELLTGKSPD 556
Query: 563 LQYPSPANRPRKVEEEETVVDLPKWVRSVVREEW-TGEVFDQELLRYKNIEEELVSMLHV 621
S VE V DL +WV+ +E E+ D LL+ + ++E++++ HV
Sbjct: 557 ----SSPGASTSVE----VPDLVRWVKKGFEQESPLSEMVDPSLLQEIHAKKEVLAVFHV 712
Query: 622 GLACVVQQPEKRPTMVDVVKMIEDI 646
L+C PE RP M V +E I
Sbjct: 713 ALSCTEGDPEVRPRMKTVSDNLERI 787
>BI264462 similar to GP|8777331|dbj receptor-like protein kinase {Arabidopsis
thaliana}, partial (26%)
Length = 620
Score = 162 bits (409), Expect = 5e-40
Identities = 93/206 (45%), Positives = 126/206 (61%), Gaps = 18/206 (8%)
Frame = +3
Query: 330 GGGG-------DSSDGTSGTDMSKLVFFDRRN-GFELEDLLRASAEMLGKGSLGTVYRAV 381
GGGG + G + +KLVFF+ + F+LEDLLRASAE+LGKGS GT Y+A+
Sbjct: 6 GGGGRGEKPKEEFGSGVQEPEKNKLVFFEGSSYNFDLEDLLRASAEVLGKGSYGTSYKAI 185
Query: 382 LDDGSTVAVKRLKDANPCARHEFEQYMDVIGKL-KHPNIVKLRAYYYAKEEKLLVYDYLS 440
L++ TV VKRLK+ + EF+Q M+++G++ +H N++ LRAYYY+K+EKLLVYDY+
Sbjct: 186 LEEAMTVVVKRLKEV-VVGKKEFDQQMEIMGRVGQHANVLPLRAYYYSKDEKLLVYDYVP 362
Query: 441 NGSLHALLHGNRGPGRIPLDWTTRISLVLGAARGLARIHT---------EYSAAKVPHGN 491
G+L LLHGNR GR PLDW +R+ + LG ARG+A IH+ +Y K P
Sbjct: 363 AGNLSTLLHGNRTGGRTPLDWDSRVKISLGTARGMAHIHSVGGPQIHTWKYQVLKCPSST 542
Query: 492 VKSSNVLLDKNGVACISDFGLSLLLN 517
K A FGL L+N
Sbjct: 543 ---------KITTAAFLIFGLXSLMN 593
>TC89046 weakly similar to GP|21740816|emb|CAD41006. OSJNBa0042L16.4 {Oryza
sativa}, partial (35%)
Length = 1399
Score = 93.6 bits (231), Expect(2) = 1e-39
Identities = 51/126 (40%), Positives = 76/126 (59%), Gaps = 5/126 (3%)
Frame = +3
Query: 356 FELEDLLRASAEM-----LGKGSLGTVYRAVLDDGSTVAVKRLKDANPCARHEFEQYMDV 410
+ L++LLRA+ +G+G G+VY G +AVKRLK A EF ++V
Sbjct: 195 YTLKELLRATNNFHQDNKIGEGGFGSVYWGQTSKGVEIAVKRLKTMTAKAEMEFAVEVEV 374
Query: 411 IGKLKHPNIVKLRAYYYAKEEKLLVYDYLSNGSLHALLHGNRGPGRIPLDWTTRISLVLG 470
+G+++H N++ LR +Y +E+L+VYDY+SN SL LHG + LDW R+S+ +G
Sbjct: 375 LGRVRHKNLLGLRGFYAGGDERLIVYDYMSNHSLLTHLHGQLASDCL-LDWPRRMSITVG 551
Query: 471 AARGLA 476
AA GLA
Sbjct: 552 AAEGLA 569
Score = 88.6 bits (218), Expect(2) = 1e-39
Identities = 54/162 (33%), Positives = 90/162 (55%), Gaps = 5/162 (3%)
Frame = +1
Query: 489 HGNVKSSNVLLDKNGVACISDFGLSLLL--NPVHATARLGG---YRAPEQTEQKRLSQQA 543
H ++K+SNVLLD A ++DFG + L+ H T R+ G Y APE ++S+
Sbjct: 607 HRDIKASNVLLDTEFQAKVADFGFAKLIPAGVSHLTTRVKGTLGYLAPEYAMWGKVSESC 786
Query: 544 DVYSFGVLLLEVLTGKAPSLQYPSPANRPRKVEEEETVVDLPKWVRSVVREEWTGEVFDQ 603
DVYSFG+LLLE+++ K P + P R D+ +WV V++ + D
Sbjct: 787 DVYSFGILLLEIISAKKPIEKLPGGIKR-----------DIVQWVTPYVQKGVFKHIADP 933
Query: 604 ELLRYKNIEEELVSMLHVGLACVVQQPEKRPTMVDVVKMIED 645
+L ++ E+L S++ + + C P+KRP+M++VV+ ++D
Sbjct: 934 KLKGNFDL-EQLKSVIMIAVRCTDSSPDKRPSMIEVVEWLKD 1056
>TC88105 similar to GP|21593085|gb|AAM65034.1 Putative protein kinase
{Arabidopsis thaliana}, partial (72%)
Length = 2379
Score = 159 bits (402), Expect = 3e-39
Identities = 112/300 (37%), Positives = 163/300 (54%), Gaps = 11/300 (3%)
Frame = +2
Query: 356 FELEDLLRASAE-----MLGKGSLGTVYRAVLDDGSTVAVKRLKDANPCARHEFEQYMDV 410
F L DL A+ + ++G+G G VY+ L +G+ VA+K+L + A EF ++
Sbjct: 689 FTLRDLELATNKFSKDNIIGEGGYGVVYQGQLINGNPVAIKKLLNNLGQAEKEFRVEVEA 868
Query: 411 IGKLKHPNIVKLRAYYYAKEEKLLVYDYLSNGSLHALLHG-NRGPGRIPLDWTTRISLVL 469
IG ++H N+V+L + +LL+Y+Y++NG+L LHG R G L W RI ++L
Sbjct: 869 IGHVRHKNLVRLLGFCIEGTHRLLIYEYVNNGNLEQWLHGAMRQYGY--LTWDARIKILL 1042
Query: 470 GAARGLARIHTEYSAAKVPHGNVKSSNVLLDKNGVACISDFGLSLLL--NPVHATARL-- 525
G A+ LA +H E KV H ++KSSN+L+D + A ISDFGL+ LL H T R+
Sbjct: 1043GTAKALAYLH-EAIEPKVVHRDIKSSNILIDDDFNAKISDFGLAKLLGAGKSHITTRVMG 1219
Query: 526 -GGYRAPEQTEQKRLSQQADVYSFGVLLLEVLTGKAPSLQYPSPANRPRKVEEEETVVDL 584
GY APE L++++DVYSFGVLLLE +TG+ P + R E V+L
Sbjct: 1220TFGYVAPEYANSGLLNEKSDVYSFGVLLLEAITGR-------DPVDYNRSAAE----VNL 1366
Query: 585 PKWVRSVVREEWTGEVFDQELLRYKNIEEELVSMLHVGLACVVQQPEKRPTMVDVVKMIE 644
W++ +V EV D + + L +L L CV EKRP M VV+M+E
Sbjct: 1367VDWLKMMVGNRHAEEVVDPN-IETRPSTSALKRVLLTALRCVDPDSEKRPKMSQVVRMLE 1543
>TC78806 similar to GP|8978273|dbj|BAA98164.1 receptor protein kinase-like
{Arabidopsis thaliana}, partial (28%)
Length = 1962
Score = 158 bits (400), Expect = 6e-39
Identities = 148/527 (28%), Positives = 241/527 (45%), Gaps = 40/527 (7%)
Frame = +2
Query: 174 SGNIPDLSSIMPNLTELNMTNNEFYGKVPNTMLNKFGDESFSGNEGLCGSKPFQVCS--- 230
SGN+ +S N ++ + +Y ++ TM++ + ++ + + S PF+ S
Sbjct: 14 SGNLTSRASSFSNFSDTS-----YYKEIEQTMMDFYRKQNIPV-DSVSLSNPFKDSSTDN 175
Query: 231 --LTENSPPS-SEPVQTVPSNPSSFPATSVIARP-------------------RSQHHKG 268
LT N PS ++ + ++F ++ + +P S+ K
Sbjct: 176 FQLTLNIFPSQTDRFNATGVSTAAFALSNQLYKPPEFFTPYAFIGVNYKHLGGESKGSKS 355
Query: 269 LSPGVIVAIVVAICVAL-LVVTSFVVAHCCARGRGVNSNSLMGSEAGKRKSYGSEKKVYN 327
GVIV VVA+ V L L + + A R R SN + E N
Sbjct: 356 SHTGVIVGAVVAVLVLLVLAILIGIYAIRQKRARSSESNPFVNWEQ-------------N 496
Query: 328 SNGGGGDSSDGTSGTDMSKLVFFDRRNGFELEDLLRASAEMLGKGSLGTVYRAVLDDGST 387
+N G G ++ + N F A A +G G G VY+ L G
Sbjct: 497 NNSGAAPQLKGARWFSFDEMRKYT--NNF-------AEANTIGSGGYGQVYQGALPTGEL 649
Query: 388 VAVKRLKDANPCARHEFEQYMDVIGKLKHPNIVKLRAYYYAKEEKLLVYDYLSNGSLHAL 447
VA+KR + EF+ ++++ ++ H N+V L + Y K E++LVY+Y+ NG+L
Sbjct: 650 VAIKRAGKESMQGAVEFKTEIELLSRVHHKNLVSLVGFCYEKGEQMLVYEYVPNGTLLDS 829
Query: 448 LHGNRGPGRIPLDWTTRISLVLGAARGLARIHTEYSAAKVPHGNVKSSNVLLDKNGVACI 507
L G I +DW R+ + LGAARGL +H E + + H ++KSSN+LLD + +A +
Sbjct: 830 LSRKSG---IWMDWIRRLKVTLGAARGLTYLH-ELADPPIIHRDIKSSNILLDNHLIAKV 997
Query: 508 SDFGLSLLL---NPVHATARLG---GYRAPEQTEQKRLSQQADVYSFGVLLLEVLTGKAP 561
+DFGLS LL H T ++ GY PE ++L++++DVYSFGVL+LE+ T + P
Sbjct: 998 ADFGLSKLLVDSERGHVTTQVKGTMGYLDPEYYMTQQLTEKSDVYSFGVLMLELATSRKP 1177
Query: 562 SLQYPSPANRPRKVEEEETVVDLPKWVRSVVREEWTGEVFDQELLRYKNIEEELVSMLHV 621
Q V E V+D K + ++ + DQ LL+ + L + +
Sbjct: 1178IEQ------GKYIVREVMRVMDTSKELYNL------HSILDQSLLKGTR-PKGLERYVEL 1318
Query: 622 GLACVVQQPEKRPTMVDVVKMIEDI--------RVEQSPLCEDYDES 660
L CV + +RP+M +V K IE I E + E+Y+E+
Sbjct: 1319ALRCVKEYAAERPSMAEVAKEIESIIELVGVNPNSESASTTENYEEA 1459
>TC92229 similar to PIR|B86369|B86369 hypothetical protein AAC98010.1
[imported] - Arabidopsis thaliana, partial (44%)
Length = 1347
Score = 157 bits (398), Expect = 1e-38
Identities = 99/290 (34%), Positives = 165/290 (56%), Gaps = 9/290 (3%)
Frame = +2
Query: 364 ASAEMLGKGSLGTVYRAVLDDGSTVAVKRLKDANPCARHEFEQYMDVIGKLKHPNIVKLR 423
+S ++G+G G VY+A++ DG A+K LK + EF +D I ++ H ++V L
Sbjct: 197 SSENVIGEGGFGRVYKALMPDGRVGALKLLKAGSGQGEREFRAEVDTISRVHHRHLVSLI 376
Query: 424 AYYYAKEEKLLVYDYLSNGSLHALLHGNRGPGRIPLDWTTRISLVLGAARGLARIHTEYS 483
Y A+++++L+Y+++ NG+L LH ++ LDW R+ + +GAARGLA +H E
Sbjct: 377 GYCIAEQQRVLIYEFVPNGNLDQHLHESQWN---VLDWPKRMKIAIGAARGLAYLH-EGC 544
Query: 484 AAKVPHGNVKSSNVLLDKNGVACISDFGLSLLLNP--VHATARL---GGYRAPEQTEQKR 538
K+ H ++KSSN+LLD + A ++DFGL+ L + H + R+ GY APE +
Sbjct: 545 NPKIIHRDIKSSNILLDDSYEAQVADFGLARLTDDTNTHVSTRVMGTFGYMAPEYATSGK 724
Query: 539 LSQQADVYSFGVLLLEVLTGKAPSLQYPSPANRPRKVEEEETVVDLPKWVRSV-VREEWT 597
L+ ++DV+SFGV+LLE++TG+ P + + V +E V +W R + +R T
Sbjct: 725 LTDRSDVFSFGVVLLELVTGR-------KPVDPTQPVGDESLV----EWARPILLRAIET 871
Query: 598 G---EVFDQELLRYKNIEEELVSMLHVGLACVVQQPEKRPTMVDVVKMIE 644
G E+ D L R + I+ E+ M+ AC+ KRP MV + + ++
Sbjct: 872 GDFSELADPRLHR-QYIDSEMFRMIEAAAACIRHSAPKRPRMVQIARALD 1018
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.316 0.134 0.392
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 20,736,431
Number of Sequences: 36976
Number of extensions: 301880
Number of successful extensions: 4206
Number of sequences better than 10.0: 832
Number of HSP's better than 10.0 without gapping: 2930
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3249
length of query: 676
length of database: 9,014,727
effective HSP length: 103
effective length of query: 573
effective length of database: 5,206,199
effective search space: 2983152027
effective search space used: 2983152027
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 62 (28.5 bits)
Medicago: description of AC139525.8


