

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC139525.4 + phase: 0
(490 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
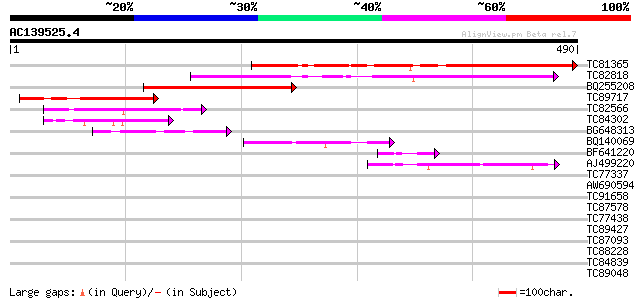
Score E
Sequences producing significant alignments: (bits) Value
TC81365 similar to GP|14579399|gb|AAK69274.1 unknown {Glycine ma... 289 1e-78
TC82818 similar to GP|20259203|gb|AAM14317.1 unknown protein {Ar... 209 2e-54
BQ255208 similar to GP|14579399|gb| unknown {Glycine max}, parti... 129 3e-30
TC89717 homologue to GP|14579399|gb|AAK69274.1 unknown {Glycine ... 108 6e-24
TC82566 weakly similar to GP|20259203|gb|AAM14317.1 unknown prot... 86 2e-17
TC84302 similar to PIR|E96797|E96797 hypothetical protein F7O12.... 64 1e-10
BG648313 similar to GP|9279604|dbj DNA-binding protein GT-1 {Ara... 45 5e-05
BQ140069 similar to GP|4031|emb|CAA Negative growth regulatory p... 43 2e-04
BF641220 weakly similar to PIR|G86203|G86 probable N-arginine di... 43 3e-04
AJ499220 similar to SP|Q09863|YAF Hypothetical protein C29E6.10c... 42 5e-04
TC77337 weakly similar to GP|4019275|gb|AAC95573.1| orf 48 {Atel... 40 0.002
AW690594 similar to GP|23498163|emb hypothetical protein {Plasmo... 40 0.002
TC91658 similar to GP|14329812|emb|CAC40753. putative nucleosome... 40 0.002
TC87578 similar to PIR|T12113|T12113 transcription factor - fava... 40 0.002
TC77438 homologue to PIR|T06807|T06807 nucleosome assembly prote... 39 0.003
TC89427 weakly similar to GP|4557063|gb|AAD22502.1| expressed pr... 38 0.008
TC87093 homologue to GP|10177817|dbj|BAB11183. gene_id:MKD15.14~... 38 0.008
TC88228 similar to GP|13605916|gb|AAK32943.1 AT4g00830/A_TM018A1... 37 0.017
TC84839 similar to GP|19386827|dbj|BAB86205. hypothetical protei... 37 0.017
TC89048 weakly similar to PIR|T46215|T46215 hypothetical protein... 37 0.022
>TC81365 similar to GP|14579399|gb|AAK69274.1 unknown {Glycine max}, partial
(38%)
Length = 920
Score = 289 bits (740), Expect = 1e-78
Identities = 159/284 (55%), Positives = 200/284 (69%), Gaps = 3/284 (1%)
Frame = +1
Query: 210 LLDSMDLSPKMKDEVRKLLNSKHLFFREMCAYHNSCGHGGVATSNVQHQVEGGSGTTTPP 269
LLD+MDLS K+K+E RKLLNSKHLFFREMCAYHNSCGHG +N+Q Q S T P
Sbjct: 1 LLDTMDLSAKLKEEARKLLNSKHLFFREMCAYHNSCGHGN---NNLQQQ----SSTEAQP 159
Query: 270 QNQPQQQHQHQHQQQNQQHCFHSSENGVGSLGVSRGEGLRMLKIGSGYVEEEDDEDEEDE 329
Q Q Q Q QQQ QQ C HSS NGVGSLG+ + +G G EEE+DEDE+DE
Sbjct: 160 QQHHHPQQQQQQQQQQQQQCLHSS-NGVGSLGMLKLKGRN-----GGGGEEEEDEDEDDE 321
Query: 330 SEDFSDEGEDESGEGC---SKGHINDQDEEENDGKPSRKRARKGGFSFPRSSSSTQLVNQ 386
ED S+E E+ESGEG SKGH EE+ + RK++RK SS +Q++ Q
Sbjct: 322 WEDDSEEEEEESGEGGGYGSKGH-----EEDENIDHLRKKSRKV------SSLPSQVMQQ 468
Query: 387 MNNEISGVFQDGGKSTWEKKQWIRNRIMQLEEQKIGYESQAFQLEKQRLKWARYSSKKER 446
+++E+ V QDG KS WEKKQW++ +++QL E+++ Y +AF++EKQRLKW ++SS KER
Sbjct: 469 LSSEVMSVLQDGVKSCWEKKQWMKKKVVQLGEEQVNYHVEAFEIEKQRLKWVKFSSNKER 648
Query: 447 EMERAKLENERRRLENERMVLLIRKKELELMHIQQQQQQQHSST 490
EMER KLENER+ LE +RMVLL+R+KELEL + QQ QQQQHSST
Sbjct: 649 EMERQKLENERKSLEIDRMVLLLRQKELELQNTQQLQQQQHSST 780
>TC82818 similar to GP|20259203|gb|AAM14317.1 unknown protein {Arabidopsis
thaliana}, partial (43%)
Length = 1176
Score = 209 bits (532), Expect = 2e-54
Identities = 122/324 (37%), Positives = 184/324 (56%), Gaps = 6/324 (1%)
Frame = +1
Query: 157 GKWKSVSRAMMEKGFYVSPQQCEDKFNDLNKRYKRVNDILGKGTACRVVENQGLLDSMD- 215
GKWKS+S+ M E+G+ VSPQQCEDKFNDLNKRYKR+ND+LG+GT+C+VVEN LLD ++
Sbjct: 1 GKWKSISKVMAERGYRVSPQQCEDKFNDLNKRYKRLNDMLGRGTSCQVVENPALLDVIEY 180
Query: 216 LSPKMKDEVRKLLNSKHLFFREMCAYHNSCGHGGVATSNVQHQVEGGSGTTTPPQNQPQQ 275
L+ K KD+VRK+LNSKHLF+ EMC+YHN N H + P
Sbjct: 181 LNEKEKDDVRKILNSKHLFYEEMCSYHN---------CNRLHL-----------PHDPAL 300
Query: 276 QHQHQHQQQNQQHCFHSSENGVGSLGVSRGEGLRMLKIGSGYVEEEDDEDEEDESEDFSD 335
Q Q +N+ H +++ + Y ++ D++D + E++D +
Sbjct: 301 QRSLQIALRNRDD--HDNDD-----------------VRRSYHDDHDEDDHDMETDDHDE 423
Query: 336 EGEDESGEGCSK---GHINDQDEEENDGKPSRKRARKG-GFSFPRSSSSTQLVNQM-NNE 390
E+ + G S+ G + + G+ G F+ S QM +
Sbjct: 424 FEENYASHGDSRVIFGGLGGTPKRLRQGQGHEDATTFGNSFNCQDYHKSPYPHGQMVQPD 603
Query: 391 ISGVFQDGGKSTWEKKQWIRNRIMQLEEQKIGYESQAFQLEKQRLKWARYSSKKEREMER 450
+ + K+ W +KQWI +R +QLEEQK+ + + +LEKQ+ KW R+S KK+RE+E+
Sbjct: 604 GNHALPENMKAAWLQKQWIESRSVQLEEQKLQIQVEMMELEKQKFKWERFSKKKDRELEK 783
Query: 451 AKLENERRRLENERMVLLIRKKEL 474
KLEN+R ++ENER+ L +++KE+
Sbjct: 784 FKLENDRMKIENERIALELKRKEI 855
>BQ255208 similar to GP|14579399|gb| unknown {Glycine max}, partial (16%)
Length = 416
Score = 129 bits (323), Expect = 3e-30
Identities = 76/135 (56%), Positives = 88/135 (64%), Gaps = 2/135 (1%)
Frame = +1
Query: 116 MKWTDTMVRLLIMAVYYIGDEAGSEGTDPNKKKSSGLLQKKGKWK-SVSRAMMEKGFYVS 174
MKWTD+MVRLLIMAVYYIGDEAGS +KKK++GLLQK+ + + S R ++ +
Sbjct: 4 MKWTDSMVRLLIMAVYYIGDEAGSLELSADKKKANGLLQKQREMEISFQRNDGKRVLCFT 183
Query: 175 PQQCEDKFNDLNKRYKRVNDILG-KGTACRVVENQGLLDSMDLSPKMKDEVRKLLNSKHL 233
R K VNDI K VVENQ LLD+MDL PK+K E RKLLNS H
Sbjct: 184 LTM*TQV**XXTTRXKGVNDISWLKEQLVAVVENQTLLDTMDLYPKLKQEPRKLLNSXHX 363
Query: 234 FFREMCAYHNSCGHG 248
FFREMCAYHNSC HG
Sbjct: 364 FFREMCAYHNSCXHG 408
>TC89717 homologue to GP|14579399|gb|AAK69274.1 unknown {Glycine max},
partial (5%)
Length = 648
Score = 108 bits (269), Expect = 6e-24
Identities = 60/121 (49%), Positives = 75/121 (61%), Gaps = 1/121 (0%)
Frame = +3
Query: 9 GLLGVVDNIPLHQQQQQQKQQNLPNQQQQNPHQLHHSQVVSYAPQHHDTDTNQHHHHSMK 68
G+ ++N L + ++QQN QNPH LHH Q+VSY H S+K
Sbjct: 330 GIFPNINNGFLGVENPSKQQQN-----HQNPHTLHHHQMVSY----------DQPHQSIK 464
Query: 69 HGFP-PFSSSKNKQQQQSSQMSDEDEPNFPAEESSGGDPKRKISPWQRMKWTDTMVRLLI 127
G+P P ++ NK QQ + +SDEDEP F A+E+S GDPKRK SPW RMKWTDTMVRLLI
Sbjct: 465 QGYPYPSKTNNNKPQQININLSDEDEPCFGADETSVGDPKRKGSPWHRMKWTDTMVRLLI 644
Query: 128 M 128
M
Sbjct: 645 M 647
>TC82566 weakly similar to GP|20259203|gb|AAM14317.1 unknown protein
{Arabidopsis thaliana}, partial (6%)
Length = 686
Score = 86.3 bits (212), Expect = 2e-17
Identities = 54/146 (36%), Positives = 77/146 (51%), Gaps = 5/146 (3%)
Frame = +1
Query: 30 NLPNQQQQNPHQLHHSQVVSYAPQHHDTDTNQHHHHSMKH-GFPPFSSSKNKQQQQSSQM 88
N QQ Q H HH +++ + D+NQ +K+ F + S+ M
Sbjct: 250 NRHQQQTQLAHGQHHMNMITGL----ENDSNQIGLIEVKNLNFGKGKGIASSNHDNSNDM 417
Query: 89 SDEDEPNFP----AEESSGGDPKRKISPWQRMKWTDTMVRLLIMAVYYIGDEAGSEGTDP 144
S++DE + E G +K SPWQRMKWTD +V LLI V +G++ G D
Sbjct: 418 SEDDEHGYGEDGNCENFFDGGKGKKGSPWQRMKWTDNVVGLLIAVVSCVGEDGTISGVDG 597
Query: 145 NKKKSSGLLQKKGKWKSVSRAMMEKG 170
K+K SG++QKKGKWK+VS+ M+ KG
Sbjct: 598 VKRK-SGVVQKKGKWKTVSKIMISKG 672
>TC84302 similar to PIR|E96797|E96797 hypothetical protein F7O12.4
[imported] - Arabidopsis thaliana, partial (7%)
Length = 832
Score = 63.9 bits (154), Expect = 1e-10
Identities = 44/133 (33%), Positives = 58/133 (43%), Gaps = 21/133 (15%)
Frame = +1
Query: 30 NLPNQQQQNPHQLHHSQVVSYAPQHHDTDTNQHH----HHSMKHGFPPFSSSKNKQQQQS 85
+LP Q + HQ HH + H N H H S++ GFP Q
Sbjct: 439 DLPGSIQVH-HQAHHPHTI------HQHQANPHQGLSLHSSVQDGFPLTMGPLQNCDQSM 597
Query: 86 SQM------------SDEDEPNF-----PAEESSGGDPKRKISPWQRMKWTDTMVRLLIM 128
S S+EDEP+F + G K SPWQR+KWTD M +L+I
Sbjct: 598 SMTDYGKGERGKICTSEEDEPSFMEDGFDGQHEGGRGKKGSSSPWQRVKWTDNMGKLMIT 777
Query: 129 AVYYIGDEAGSEG 141
AV YIG++ S+G
Sbjct: 778 AVSYIGEDRTSDG 816
>BG648313 similar to GP|9279604|dbj DNA-binding protein GT-1 {Arabidopsis
thaliana}, partial (53%)
Length = 815
Score = 45.4 bits (106), Expect = 5e-05
Identities = 39/122 (31%), Positives = 53/122 (42%), Gaps = 2/122 (1%)
Frame = +1
Query: 72 PPFSSSKNKQQQQSSQMSDEDEPNFPAEESSGGDPKRKI-SPWQRMK-WTDTMVRLLIMA 129
PP S + QQQ QM ESSG DP+ +I +P +R + W R LI
Sbjct: 37 PPPSQHHHHLQQQQPQM-------ILTAESSGDDPEMEIKAPKKRAETWVQDETRSLI-- 189
Query: 130 VYYIGDEAGSEGTDPNKKKSSGLLQKKGKWKSVSRAMMEKGFYVSPQQCEDKFNDLNKRY 189
G + K + L W+ +S M EKGF SP C DK+ +L K +
Sbjct: 190 ----GLRREMDSLFNTSKSNKHL------WEQISAKMREKGFDRSPTMCTDKWRNLLKEF 339
Query: 190 KR 191
K+
Sbjct: 340 KK 345
>BQ140069 similar to GP|4031|emb|CAA Negative growth regulatory protein
{Saccharomyces cerevisiae}, partial (3%)
Length = 606
Score = 43.1 bits (100), Expect = 2e-04
Identities = 35/134 (26%), Positives = 58/134 (43%), Gaps = 4/134 (2%)
Frame = +3
Query: 203 RVVENQGLLDSMDLSPKMKDEVRKLLNSKHLFFREMCAYHNSCGHGGVATSNVQHQVEGG 262
+++ N LD L K D++ L+N+ ++ RE + N + S Q ++
Sbjct: 123 KLLLNLSQLDLKILHSKKLDQID*LINNTNILLREGHFH*NG---NAQSQSQSQQPLKTM 293
Query: 263 SGTTTPPQN----QPQQQHQHQHQQQNQQHCFHSSENGVGSLGVSRGEGLRMLKIGSGYV 318
+ Q Q Q QHQHQHQQ +QQ F+ S + R+L + S
Sbjct: 294 NKPILISQEDQLLQQQHQHQHQHQQLDQQDFFYESND-------------RLLSLSSSCS 434
Query: 319 EEEDDEDEEDESED 332
+ D+EDE ++
Sbjct: 435 XSNSNSDQEDEDQE 476
>BF641220 weakly similar to PIR|G86203|G86 probable N-arginine dibasic
convertase [imported] - Arabidopsis thaliana, partial
(5%)
Length = 634
Score = 42.7 bits (99), Expect = 3e-04
Identities = 25/53 (47%), Positives = 28/53 (52%)
Frame = +2
Query: 319 EEEDDEDEEDESEDFSDEGEDESGEGCSKGHINDQDEEENDGKPSRKRARKGG 371
EEEDDEDEED+ ED DEGED D+DEE D R+GG
Sbjct: 371 EEEDDEDEEDDEED-DDEGED------------DEDEEXEDEDEXXVXGREGG 490
Score = 29.3 bits (64), Expect = 3.6
Identities = 14/28 (50%), Positives = 16/28 (57%)
Frame = +2
Query: 319 EEEDDEDEEDESEDFSDEGEDESGEGCS 346
E EDDEDEE E ED E G+G +
Sbjct: 419 EGEDDEDEEXEDEDEXXVXGREGGKGAA 502
>AJ499220 similar to SP|Q09863|YAF Hypothetical protein C29E6.10c in
chromosome I. [Fission yeast] {Schizosaccharomyces
pombe}, partial (12%)
Length = 518
Score = 42.0 bits (97), Expect = 5e-04
Identities = 48/182 (26%), Positives = 75/182 (40%), Gaps = 16/182 (8%)
Frame = -3
Query: 310 MLKIGSGYVEEEDDEDEEDESEDFSDEGEDESGEGCSKGHINDQDEEENDG----KPSRK 365
M ++ ++ E+DE D E + DE EDE D+DE E DG + +
Sbjct: 516 MERLAERRMQREEDESAMDRGE-YYDEDEDE-----------DEDEYEEDGEEDNQTEEQ 373
Query: 366 RARKGGFSFPRSSSSTQLVNQMNNEISGVFQDGGKSTWEKKQWIRNRIMQLEEQKIGYES 425
R +G F ++ +N V Q+ + E+ + NR+ + E K E
Sbjct: 372 RMEEGRRMFQIFAARMFEQRVLNAYREKVAQERQQKLLEELE-EENRLKEERELKKLKEK 196
Query: 426 QAFQLEKQRLKWARYSSKKEREMER------------AKLENERRRLENERMVLLIRKKE 473
+ + + + LK + + RE ER KLE ER+R E ER+ KKE
Sbjct: 195 ERKKAKNRALKQQKEEERARREAERLAEEKAQRAEREKKLEEERKRREEERL-----KKE 31
Query: 474 LE 475
E
Sbjct: 30 AE 25
>TC77337 weakly similar to GP|4019275|gb|AAC95573.1| orf 48 {Ateline
herpesvirus 3}, partial (8%)
Length = 986
Score = 40.4 bits (93), Expect = 0.002
Identities = 17/50 (34%), Positives = 30/50 (60%)
Frame = +3
Query: 319 EEEDDEDEEDESEDFSDEGEDESGEGCSKGHINDQDEEENDGKPSRKRAR 368
+E+DDEDE+D+ E+ E ++E G ++DE+E +P +KR +
Sbjct: 627 DEDDDEDEDDDDEEEGGEEDEEEGVDEEDNEEEEEDEDEEALQPPKKRKK 776
Score = 38.5 bits (88), Expect = 0.006
Identities = 19/42 (45%), Positives = 26/42 (61%), Gaps = 1/42 (2%)
Frame = +3
Query: 319 EEEDDEDEEDESEDFS-DEGEDESGEGCSKGHINDQDEEEND 359
EEEDDED +D+ ED DE +D+ EG + DEE+N+
Sbjct: 594 EEEDDEDGDDQDEDDDEDEDDDDEEEGGEEDEEEGVDEEDNE 719
Score = 37.0 bits (84), Expect = 0.017
Identities = 16/41 (39%), Positives = 28/41 (68%)
Frame = +3
Query: 319 EEEDDEDEEDESEDFSDEGEDESGEGCSKGHINDQDEEEND 359
E+ DD+DE+D+ ED D+ E+E GE + ++++D EE +
Sbjct: 609 EDGDDQDEDDD-EDEDDDDEEEGGEEDEEEGVDEEDNEEEE 728
Score = 31.6 bits (70), Expect = 0.72
Identities = 20/94 (21%), Positives = 39/94 (41%)
Frame = +3
Query: 266 TTPPQNQPQQQHQHQHQQQNQQHCFHSSENGVGSLGVSRGEGLRMLKIGSGYVEEEDDED 325
TT ++ Q Q + + + E+ G GE + G GY +++
Sbjct: 363 TTKGDSKTQSQDKQHDAEGEDGNDDEDEEDDDGDGAFGEGEDELSSEDGGGYGNNSNNKS 542
Query: 326 EEDESEDFSDEGEDESGEGCSKGHINDQDEEEND 359
++ + G DE+GE +DQDE++++
Sbjct: 543 NSKKAPEGGAGGADENGEEEDDEDGDDQDEDDDE 644
Score = 30.8 bits (68), Expect = 1.2
Identities = 15/51 (29%), Positives = 28/51 (54%)
Frame = +3
Query: 321 EDDEDEEDESEDFSDEGEDESGEGCSKGHINDQDEEENDGKPSRKRARKGG 371
+D+++E+D+ + EGEDE G+ N+ + + N K+A +GG
Sbjct: 432 DDEDEEDDDGDGAFGEGEDELSSEDGGGYGNNSNNKSNS-----KKAPEGG 569
>AW690594 similar to GP|23498163|emb hypothetical protein {Plasmodium
falciparum 3D7}, partial (10%)
Length = 633
Score = 40.0 bits (92), Expect = 0.002
Identities = 17/44 (38%), Positives = 29/44 (65%)
Frame = +3
Query: 316 GYVEEEDDEDEEDESEDFSDEGEDESGEGCSKGHINDQDEEEND 359
G VEEE+DE +E+E ++ +E E+E E H ++++EEE +
Sbjct: 72 GEVEEEEDEHDEEEEDEHDEEEEEEEEEEEEDEHDDEEEEEEEE 203
Score = 37.0 bits (84), Expect = 0.017
Identities = 16/41 (39%), Positives = 27/41 (65%)
Frame = +3
Query: 319 EEEDDEDEEDESEDFSDEGEDESGEGCSKGHINDQDEEEND 359
EEE+DE +E+E E+ +E EDE + + +++EEE+D
Sbjct: 105 EEEEDEHDEEEEEEEEEEEEDEHDDEEEEEEEEEEEEEEDD 227
Score = 34.3 bits (77), Expect = 0.11
Identities = 14/39 (35%), Positives = 25/39 (63%)
Frame = +3
Query: 319 EEEDDEDEEDESEDFSDEGEDESGEGCSKGHINDQDEEE 357
+E D+E+EE+E E+ DE +DE E + ++D++E
Sbjct: 117 DEHDEEEEEEEEEEEEDEHDDEEEEEEEEEEEEEEDDDE 233
Score = 33.9 bits (76), Expect = 0.15
Identities = 17/88 (19%), Positives = 37/88 (41%)
Frame = +3
Query: 256 QHQVEGGSGTTTPPQNQPQQQHQHQHQQQNQQHCFHSSENGVGSLGVSRGEGLRMLKIGS 315
+H E G +++ ++ + +H ++ ++ E+
Sbjct: 48 EHNEEEDEGEVEEEEDEHDEEEEDEHDEEEEEEEEEEEED-------------------- 167
Query: 316 GYVEEEDDEDEEDESEDFSDEGEDESGE 343
E +D+E+EE+E E+ +E +DE GE
Sbjct: 168 ---EHDDEEEEEEEEEEEEEEDDDEEGE 242
Score = 32.7 bits (73), Expect = 0.32
Identities = 14/42 (33%), Positives = 26/42 (61%)
Frame = +3
Query: 318 VEEEDDEDEEDESEDFSDEGEDESGEGCSKGHINDQDEEEND 359
VEE ++E++E E E+ DE ++E + + +++EEE D
Sbjct: 42 VEEHNEEEDEGEVEEEEDEHDEEEEDEHDEEEEEEEEEEEED 167
Score = 32.7 bits (73), Expect = 0.32
Identities = 12/22 (54%), Positives = 19/22 (85%)
Frame = +3
Query: 319 EEEDDEDEEDESEDFSDEGEDE 340
EEE++E+EE+E ED +EGE++
Sbjct: 183 EEEEEEEEEEEEEDDDEEGEED 248
Score = 31.6 bits (70), Expect = 0.72
Identities = 13/41 (31%), Positives = 25/41 (60%)
Frame = +3
Query: 319 EEEDDEDEEDESEDFSDEGEDESGEGCSKGHINDQDEEEND 359
EEED+ + E+E ++ +E EDE E + ++++E +D
Sbjct: 57 EEEDEGEVEEEEDEHDEEEEDEHDEEEEEEEEEEEEDEHDD 179
Score = 28.9 bits (63), Expect = 4.7
Identities = 11/36 (30%), Positives = 23/36 (63%)
Frame = +3
Query: 319 EEEDDEDEEDESEDFSDEGEDESGEGCSKGHINDQD 354
EEE++++ +DE E+ +E E+E + +G ++ D
Sbjct: 150 EEEEEDEHDDEEEEEEEEEEEEEEDDDEEGEEDEID 257
Score = 28.5 bits (62), Expect = 6.1
Identities = 18/46 (39%), Positives = 25/46 (54%)
Frame = +3
Query: 319 EEEDDEDEEDESEDFSDEGEDESGEGCSKGHINDQDEEENDGKPSR 364
EEE++E EEDE +D +E E+E E D DEE + + R
Sbjct: 141 EEEEEE-EEDEHDDEEEEEEEEEEE-----EEEDDDEEGEEDEIDR 260
>TC91658 similar to GP|14329812|emb|CAC40753. putative nucleosome assembly
protein 1 {Atropa belladonna}, partial (46%)
Length = 583
Score = 40.0 bits (92), Expect = 0.002
Identities = 18/63 (28%), Positives = 34/63 (53%)
Frame = +2
Query: 320 EEDDEDEEDESEDFSDEGEDESGEGCSKGHINDQDEEENDGKPSRKRARKGGFSFPRSSS 379
+EDDED ++E +D D+ +D+ D+D+EE D + ++++G + P
Sbjct: 86 DEDDEDGDEEDDDDDDDDDDDE---------EDEDDEEEDEGKGKSKSKRGSKAKPGKDQ 238
Query: 380 STQ 382
ST+
Sbjct: 239 STE 247
Score = 35.8 bits (81), Expect = 0.038
Identities = 15/49 (30%), Positives = 28/49 (56%)
Frame = +2
Query: 316 GYVEEEDDEDEEDESEDFSDEGEDESGEGCSKGHINDQDEEENDGKPSR 364
G E++DD+D++D+ E+ D+ E++ G+G SK + + D R
Sbjct: 104 GDEEDDDDDDDDDDDEEDEDDEEEDEGKGKSKSKRGSKAKPGKDQSTER 250
>TC87578 similar to PIR|T12113|T12113 transcription factor - fava bean,
partial (49%)
Length = 1021
Score = 40.0 bits (92), Expect = 0.002
Identities = 39/167 (23%), Positives = 70/167 (41%), Gaps = 19/167 (11%)
Frame = +2
Query: 319 EEEDDEDEEDESEDFSDEGEDESGEGCSKGHINDQDEEENDGKPSRKRARKGGFSFPRSS 378
E+E DE++ED + DEG G + +E+ P++K +K S +S
Sbjct: 524 EDESDEEDEDFVAEKDDEGSPTDDSGADDSDASQSGDEKE--IPAKKEPKKDLSSKASAS 697
Query: 379 SSTQLVNQMNNEISGVFQDGGKSTWEKKQWIRNRIMQLEEQKIGYESQAF--QLEKQRLK 436
+ST + + + +DG K +KK+ K G F Q+E++ +K
Sbjct: 698 TSTSTSKKKSKDAD---EDGKKKKQKKKK-------DPNAPKRGMSGFMFFSQMERENIK 847
Query: 437 WA-----------------RYSSKKEREMERAKLENERRRLENERMV 466
A + S +E+E AK +++R E+E++V
Sbjct: 848 KANPGISFTDVAKLLGENWKKMSAEEKEPYEAKARVDKKRYEDEKIV 988
>TC77438 homologue to PIR|T06807|T06807 nucleosome assembly protein 1 - garden
pea, partial (93%)
Length = 1607
Score = 39.3 bits (90), Expect = 0.003
Identities = 20/58 (34%), Positives = 34/58 (58%)
Frame = +2
Query: 316 GYVEEEDDEDEEDESEDFSDEGEDESGEGCSKGHINDQDEEENDGKPSRKRARKGGFS 373
G +++EDD++++D +D DE EDE + +D+DEEE K + A+K G +
Sbjct: 1028 GDLDDEDDDEDDDAEDDDEDEDEDEDED-------DDEDEEETKTK-KKSSAKKSGIA 1177
Score = 31.2 bits (69), Expect = 0.94
Identities = 19/59 (32%), Positives = 28/59 (47%)
Frame = +2
Query: 319 EEEDDEDEEDESEDFSDEGEDESGEGCSKGHINDQDEEENDGKPSRKRARKGGFSFPRS 377
EE D D+ED+ ED E +DE D+DE+E+D + + K S +S
Sbjct: 1019 EEFGDLDDEDDDEDDDAEDDDED---------EDEDEDEDDDEDEEETKTKKKSSAKKS 1168
>TC89427 weakly similar to GP|4557063|gb|AAD22502.1| expressed protein
{Arabidopsis thaliana}, partial (50%)
Length = 753
Score = 38.1 bits (87), Expect = 0.008
Identities = 19/57 (33%), Positives = 35/57 (61%), Gaps = 3/57 (5%)
Frame = +1
Query: 313 IGSGYVEEEDDEDEEDESEDFSDEGEDESGEGCSKGHINDQDEEENDGK---PSRKR 366
+ +G +DD++++D+ +D +D+GEDE D+DEEE+D + PS+K+
Sbjct: 424 VPNGAGGSDDDDEDDDDDDDDNDDGEDE-----------DEDEEEDDDEDQPPSKKK 561
Score = 33.1 bits (74), Expect = 0.25
Identities = 15/42 (35%), Positives = 23/42 (54%)
Frame = +1
Query: 318 VEEEDDEDEEDESEDFSDEGEDESGEGCSKGHINDQDEEEND 359
V +EDD+++ED S D DE D + G D++E+D
Sbjct: 343 VNDEDDDNDEDFSGDEDDEDADPEDDPVPNGAGGSDDDDEDD 468
Score = 31.6 bits (70), Expect = 0.72
Identities = 15/40 (37%), Positives = 27/40 (67%)
Frame = +1
Query: 320 EEDDEDEEDESEDFSDEGEDESGEGCSKGHINDQDEEEND 359
E +D+D+ED+ +D +DE +D++ E S D+D+E+ D
Sbjct: 304 ETEDDDDEDDDDDVNDE-DDDNDEDFS----GDEDDEDAD 408
Score = 28.9 bits (63), Expect = 4.7
Identities = 17/56 (30%), Positives = 26/56 (46%), Gaps = 4/56 (7%)
Frame = +1
Query: 308 LRMLKIGSGYVEEEDD----EDEEDESEDFSDEGEDESGEGCSKGHINDQDEEEND 359
L L G+ EE D ED++DE +D ED+ + G +D+D + D
Sbjct: 250 LDQLSKGNHTCEENKDGSETEDDDDEDDDDDVNDEDDDNDEDFSGDEDDEDADPED 417
>TC87093 homologue to GP|10177817|dbj|BAB11183. gene_id:MKD15.14~unknown
protein {Arabidopsis thaliana}, partial (49%)
Length = 1313
Score = 38.1 bits (87), Expect = 0.008
Identities = 20/40 (50%), Positives = 23/40 (57%)
Frame = +3
Query: 7 PSGLLGVVDNIPLHQQQQQQKQQNLPNQQQQNPHQLHHSQ 46
P G N H QQQQQ+QQ+ QQQQ+ HQ HH Q
Sbjct: 576 PEGFSVSQHNSLFHHQQQQQQQQH-QQQQQQHHHQQHHQQ 692
Score = 32.7 bits (73), Expect = 0.32
Identities = 21/64 (32%), Positives = 26/64 (39%), Gaps = 8/64 (12%)
Frame = +3
Query: 270 QNQPQQQHQHQHQQQNQQHCFHSSENGVGSLGVSRGEG--------LRMLKIGSGYVEEE 321
Q Q QQQHQ Q QQ + Q + +G +R L L G+G
Sbjct: 624 QQQQQQQHQQQQQQHHHQQHHQQQQGSMGEASAARLGNYLPGHLNLLASLSGGNGNSGRG 803
Query: 322 DDED 325
DDED
Sbjct: 804 DDED 815
>TC88228 similar to GP|13605916|gb|AAK32943.1 AT4g00830/A_TM018A10_14
{Arabidopsis thaliana}, partial (67%)
Length = 1712
Score = 37.0 bits (84), Expect = 0.017
Identities = 21/74 (28%), Positives = 31/74 (41%)
Frame = +1
Query: 317 YVEEEDDEDEEDESEDFSDEGEDESGEGCSKGHINDQDEEENDGKPSRKRARKGGFSFPR 376
Y+EE DD+ EE +D D GEDE+ EG + H ++ E K K
Sbjct: 205 YMEEMDDDVEEQIDDDGVDGGEDENAEGSVEEHEYEETAAEAGQKDQFPEGEKSDHGAEE 384
Query: 377 SSSSTQLVNQMNNE 390
L+++ E
Sbjct: 385 DELKPALIDEEERE 426
>TC84839 similar to GP|19386827|dbj|BAB86205. hypothetical protein~similar
to Arabidopsis thaliana chromosome 5 T6I14_10, partial
(20%)
Length = 819
Score = 37.0 bits (84), Expect = 0.017
Identities = 28/86 (32%), Positives = 33/86 (37%)
Frame = +1
Query: 21 QQQQQQKQQNLPNQQQQNPHQLHHSQVVSYAPQHHDTDTNQHHHHSMKHGFPPFSSSKNK 80
QQQQQQ+QQ +QQQQ P HH Q Q + H PP
Sbjct: 202 QQQQQQQQQYQQHQQQQQPPHFHHQQ------QQQQSGPGPDFHRGPP---PPMPQQPPP 354
Query: 81 QQQQSSQMSDEDEPNFPAEESSGGDP 106
+Q S S N +E GG P
Sbjct: 355 MMRQPSASS----TNLGSEFLPGGPP 420
Score = 33.9 bits (76), Expect = 0.15
Identities = 14/26 (53%), Positives = 17/26 (64%)
Frame = +1
Query: 269 PQNQPQQQHQHQHQQQNQQHCFHSSE 294
PQ+Q QQHQ Q QQQ QQ+ H +
Sbjct: 172 PQHQQHQQHQQQQQQQQQQYQQHQQQ 249
Score = 29.6 bits (65), Expect = 2.7
Identities = 13/25 (52%), Positives = 14/25 (56%)
Frame = +1
Query: 270 QNQPQQQHQHQHQQQNQQHCFHSSE 294
Q Q QQQ QHQQQ Q FH +
Sbjct: 205 QQQQQQQQYQQHQQQQQPPHFHHQQ 279
Score = 28.1 bits (61), Expect = 8.0
Identities = 13/27 (48%), Positives = 15/27 (55%), Gaps = 1/27 (3%)
Frame = +1
Query: 269 PQNQPQQQHQ-HQHQQQNQQHCFHSSE 294
P N QQHQ HQ QQQ QQ + +
Sbjct: 163 PHNPQHQQHQQHQQQQQQQQQQYQQHQ 243
>TC89048 weakly similar to PIR|T46215|T46215 hypothetical protein T8P19.220
- Arabidopsis thaliana, partial (35%)
Length = 1329
Score = 36.6 bits (83), Expect = 0.022
Identities = 31/160 (19%), Positives = 73/160 (45%), Gaps = 12/160 (7%)
Frame = +2
Query: 320 EEDDEDEEDESEDFSDEGEDESGEGCSKGHINDQDEEENDGKPSRKRARKGGFSFPRSSS 379
++DDEDE + +D D+ E+++GE KG + E+++ P+ R + + R S
Sbjct: 461 DKDDEDEGGDEDDADDDEEEDAGEE-KKGVKKESSEKKSPVTPTSDRPTRERKTVERYSE 637
Query: 380 STQLVNQMNNEISGVFQDGGKSTWEKKQWIRNRIMQLEEQK------------IGYESQA 427
+ ++ G+ + G+ T K I N +L ++K G +++
Sbjct: 638 PSPSKFGRSSSSKGLIIEKGRGTQLKD--IPNVAFKLSKRKPDDNLHMLHSLLFGKKTKV 811
Query: 428 FQLEKQRLKWARYSSKKEREMERAKLENERRRLENERMVL 467
L++ +++ Y + E +R +++ + E++++
Sbjct: 812 NNLKRNIGQFSGYVWAENEEKQRTRVKERIDKCVKEKLIV 931
Score = 28.9 bits (63), Expect = 4.7
Identities = 16/53 (30%), Positives = 25/53 (46%)
Frame = +2
Query: 318 VEEEDDEDEEDESEDFSDEGEDESGEGCSKGHINDQDEEENDGKPSRKRARKG 370
V E D++ E D ED + EDE G D+D+ ++D + +KG
Sbjct: 413 VAESDEKKEIDGHEDSDKDDEDEGG---------DEDDADDDEEEDAGEEKKG 544
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.310 0.128 0.368
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 16,306,547
Number of Sequences: 36976
Number of extensions: 274993
Number of successful extensions: 5766
Number of sequences better than 10.0: 359
Number of HSP's better than 10.0 without gapping: 3585
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 4886
length of query: 490
length of database: 9,014,727
effective HSP length: 100
effective length of query: 390
effective length of database: 5,317,127
effective search space: 2073679530
effective search space used: 2073679530
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 60 (27.7 bits)
Medicago: description of AC139525.4


