

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC139525.10 + phase: 0
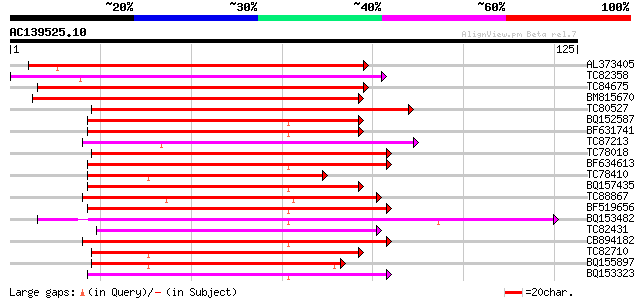
(125 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
AL373405 similar to PIR|F86331|F863 hypothetical protein AAG1254... 93 3e-20
TC82358 weakly similar to PIR|F84787|F84787 probable auxin-induc... 74 1e-14
TC84675 similar to GP|21553798|gb|AAM62891.1 unknown {Arabidopsi... 71 1e-13
BM815670 similar to GP|20149050|gb| auxin-induced SAUR-like prot... 70 3e-13
TC80527 similar to GP|11994425|dbj|BAB02427. auxin-regulated pro... 63 3e-11
BQ152587 similar to SP|P33080|AX10_S Auxin-induced protein X10A.... 61 1e-10
BF631741 similar to SP|P32295|ARG7_P Indole-3-acetic acid induce... 61 1e-10
TC87213 similar to PIR|G84887|G84887 probable auxin-regulated pr... 60 2e-10
TC78018 similar to PIR|T04499|T04499 auxin-induced protein homol... 60 2e-10
BF634613 similar to SP|P33083|AX6B_S Auxin-induced protein 6B. [... 60 2e-10
TC78410 similar to PIR|A84906|A84906 probable auxin-regulated pr... 60 2e-10
BQ157435 similar to SP|P33080|AX10_S Auxin-induced protein X10A.... 59 4e-10
TC88867 weakly similar to GP|9758890|dbj|BAB09466.1 auxin-induce... 59 6e-10
BF519656 similar to SP|P32295|ARG7_P Indole-3-acetic acid induce... 58 1e-09
BQ153482 similar to PIR|T06083|T0608 probable auxin-induced prot... 57 2e-09
TC82431 weakly similar to GP|8777399|dbj|BAA96989.1 emb|CAB86483... 56 3e-09
CB894182 similar to SP|P33083|AX6B_S Auxin-induced protein 6B. [... 56 3e-09
TC82710 similar to PIR|A84906|A84906 probable auxin-regulated pr... 56 3e-09
BQ155897 similar to PIR|T12211|T1221 auxin-induced protein - com... 56 3e-09
BQ153323 similar to PIR|T06083|T0608 probable auxin-induced prot... 56 4e-09
>AL373405 similar to PIR|F86331|F863 hypothetical protein AAG12548.1
[imported] - Arabidopsis thaliana, partial (64%)
Length = 455
Score = 92.8 bits (229), Expect = 3e-20
Identities = 45/77 (58%), Positives = 53/77 (68%), Gaps = 2/77 (2%)
Frame = +2
Query: 5 WRKNA--CSGKKLPSDVPRGHLAVTVGETNRRFVIRADYLNHPVLQELLDQAYEGYGFNK 62
WR A S + PSDVP GH+AV VG RFV+RA YLNHP+ Q+LL QA E YGF+
Sbjct: 107 WRNKARISSANRAPSDVPSGHVAVCVGANYTRFVVRATYLNHPIFQKLLVQAEEEYGFSN 286
Query: 63 SGPLSIPCDEFLFEDIL 79
GPL+IPCDE FE+ L
Sbjct: 287 HGPLTIPCDEEFFEEAL 337
>TC82358 weakly similar to PIR|F84787|F84787 probable auxin-induced protein
[imported] - Arabidopsis thaliana, partial (50%)
Length = 546
Score = 74.3 bits (181), Expect = 1e-14
Identities = 38/89 (42%), Positives = 52/89 (57%), Gaps = 6/89 (6%)
Frame = +3
Query: 1 MACMWRKNACSGKK------LPSDVPRGHLAVTVGETNRRFVIRADYLNHPVLQELLDQA 54
+ C R + CS + +P DVP+GHL V VGE +RFVI+ LNHP + LLD A
Sbjct: 99 LKCAIRASFCSSSQQKSNLHIPKDVPKGHLVVYVGEDCKRFVIKVGTLNHPPFKALLDHA 278
Query: 55 YEGYGFNKSGPLSIPCDEFLFEDILLSLG 83
+ +GF L IPC+E +F +IL + G
Sbjct: 279 EDAFGFTNGSKLLIPCNENVFLNILHNAG 365
>TC84675 similar to GP|21553798|gb|AAM62891.1 unknown {Arabidopsis
thaliana}, partial (46%)
Length = 599
Score = 70.9 bits (172), Expect = 1e-13
Identities = 32/73 (43%), Positives = 51/73 (69%)
Frame = +1
Query: 7 KNACSGKKLPSDVPRGHLAVTVGETNRRFVIRADYLNHPVLQELLDQAYEGYGFNKSGPL 66
+++C+ K P DVP+G+LAV VG RRF+I YL+H + + LL++A + +GFN+ G L
Sbjct: 115 EDSCNSPKAPHDVPKGYLAVYVGPELRRFIIPTSYLSHSLFKMLLEKAADEFGFNQCGGL 294
Query: 67 SIPCDEFLFEDIL 79
+IPC+ F+ +L
Sbjct: 295 TIPCEIETFKYLL 333
>BM815670 similar to GP|20149050|gb| auxin-induced SAUR-like protein
{Capsicum annuum}, partial (80%)
Length = 611
Score = 69.7 bits (169), Expect = 3e-13
Identities = 31/73 (42%), Positives = 45/73 (61%)
Frame = +3
Query: 6 RKNACSGKKLPSDVPRGHLAVTVGETNRRFVIRADYLNHPVLQELLDQAYEGYGFNKSGP 65
++ + + + LP DVP+GH V VGE R++I +L HP Q LL +A + +GFN
Sbjct: 33 KRQSYNEEGLPEDVPKGHFVVYVGENRTRYIIPISWLAHPQFQSLLQRAEDEFGFNHDMG 212
Query: 66 LSIPCDEFLFEDI 78
L+IPCDE FE +
Sbjct: 213 LTIPCDEVFFESL 251
>TC80527 similar to GP|11994425|dbj|BAB02427. auxin-regulated protein-like
{Arabidopsis thaliana}, partial (62%)
Length = 632
Score = 62.8 bits (151), Expect = 3e-11
Identities = 29/71 (40%), Positives = 45/71 (62%)
Frame = +1
Query: 19 VPRGHLAVTVGETNRRFVIRADYLNHPVLQELLDQAYEGYGFNKSGPLSIPCDEFLFEDI 78
VP GH+ + VG+ RFV+ A+ LNHPV +LL+++ + YG+ + G L +PC +FE +
Sbjct: 259 VPEGHVPIYVGDEMERFVVCAELLNHPVFIKLLNESAQEYGYEQKGVLRLPCHVLVFERV 438
Query: 79 LLSLGGGTVAR 89
L +L G R
Sbjct: 439 LEALKLGLDTR 471
>BQ152587 similar to SP|P33080|AX10_S Auxin-induced protein X10A. [Soybean]
{Glycine max}, partial (92%)
Length = 475
Score = 61.2 bits (147), Expect = 1e-10
Identities = 33/62 (53%), Positives = 43/62 (69%), Gaps = 1/62 (1%)
Frame = +2
Query: 18 DVPRGHLAVTVGETNRRFVIRADYLNHPVLQELLDQAYEGYGF-NKSGPLSIPCDEFLFE 76
DVP+G+LAV VGE +RFVI YL+ QELL+QA E +G+ + G L+IPC E +F
Sbjct: 155 DVPKGYLAVYVGEEMKRFVIPISYLSQSSFQELLNQAEEQFGYDHPMGGLTIPCREDVFL 334
Query: 77 DI 78
DI
Sbjct: 335 DI 340
>BF631741 similar to SP|P32295|ARG7_P Indole-3-acetic acid induced protein
ARG7. [Mung bean Vigna radiata] {Phaseolus aureus},
partial (78%)
Length = 450
Score = 60.8 bits (146), Expect = 1e-10
Identities = 32/62 (51%), Positives = 44/62 (70%), Gaps = 1/62 (1%)
Frame = +1
Query: 18 DVPRGHLAVTVGETNRRFVIRADYLNHPVLQELLDQAYEGYGF-NKSGPLSIPCDEFLFE 76
+VP+G LAV VGE +RFVI YLN P+ Q+LL+QA E + + + +G L+IPC E +F
Sbjct: 25 EVPKGCLAVYVGEEMKRFVIPISYLNQPLFQDLLNQAEEQFEYDHPTGGLTIPCREDMFL 204
Query: 77 DI 78
DI
Sbjct: 205DI 210
>TC87213 similar to PIR|G84887|G84887 probable auxin-regulated protein
[imported] - Arabidopsis thaliana, partial (44%)
Length = 809
Score = 60.5 bits (145), Expect = 2e-10
Identities = 32/77 (41%), Positives = 45/77 (57%), Gaps = 3/77 (3%)
Frame = +1
Query: 17 SDVPRGHLAVTVGETN---RRFVIRADYLNHPVLQELLDQAYEGYGFNKSGPLSIPCDEF 73
S VP+GHLAV VG+ + RR ++ Y NHP+ ELL +A + YGF G ++IPC
Sbjct: 250 SPVPKGHLAVYVGQKDGEFRRVLVPVVYFNHPLFGELLKEAEKEYGFCHQGGITIPCRVT 429
Query: 74 LFEDILLSLGGGTVARR 90
FE + + G+ RR
Sbjct: 430 EFERVKTRIASGSDTRR 480
>TC78018 similar to PIR|T04499|T04499 auxin-induced protein homolog
F8F16.140 - Arabidopsis thaliana, partial (58%)
Length = 803
Score = 60.5 bits (145), Expect = 2e-10
Identities = 31/66 (46%), Positives = 41/66 (61%)
Frame = +2
Query: 19 VPRGHLAVTVGETNRRFVIRADYLNHPVLQELLDQAYEGYGFNKSGPLSIPCDEFLFEDI 78
VP+G+LAV VG RFVI +YL H LL +A E +GF ++G L IPC+ +FE I
Sbjct: 350 VPKGYLAVCVGVDLNRFVIPTEYLAHQAFHILLREAEEEFGFEQTGVLRIPCEVSVFESI 529
Query: 79 LLSLGG 84
L + G
Sbjct: 530 LKMVEG 547
>BF634613 similar to SP|P33083|AX6B_S Auxin-induced protein 6B. [Soybean]
{Glycine max}, partial (82%)
Length = 368
Score = 60.1 bits (144), Expect = 2e-10
Identities = 31/68 (45%), Positives = 45/68 (65%), Gaps = 1/68 (1%)
Frame = +3
Query: 18 DVPRGHLAVTVGETNRRFVIRADYLNHPVLQELLDQAYEGYGF-NKSGPLSIPCDEFLFE 76
+V +G++AV VGE RFV+ YLN P Q+LL Q+ E +G+ + G L+IPC E +F+
Sbjct: 27 EVRKGYVAVYVGEKLVRFVVPVSYLNQPSFQDLLSQSEEEFGYDHPMGGLTIPCTEDVFQ 206
Query: 77 DILLSLGG 84
I+ SL G
Sbjct: 207HIISSLNG 230
>TC78410 similar to PIR|A84906|A84906 probable auxin-regulated protein
[imported] - Arabidopsis thaliana, partial (61%)
Length = 1073
Score = 60.1 bits (144), Expect = 2e-10
Identities = 28/55 (50%), Positives = 38/55 (68%), Gaps = 2/55 (3%)
Frame = +1
Query: 18 DVPRGHLAVTVG--ETNRRFVIRADYLNHPVLQELLDQAYEGYGFNKSGPLSIPC 70
D+P+G LAV VG E ++FVI Y+NHP+ +LL +A E YGF+ GP+ IPC
Sbjct: 625 DIPKGCLAVMVGQGEEQQKFVIPVIYINHPLFMQLLKEAEEEYGFDHKGPIIIPC 789
>BQ157435 similar to SP|P33080|AX10_S Auxin-induced protein X10A. [Soybean]
{Glycine max}, partial (89%)
Length = 463
Score = 59.3 bits (142), Expect = 4e-10
Identities = 32/62 (51%), Positives = 41/62 (65%), Gaps = 1/62 (1%)
Frame = +3
Query: 18 DVPRGHLAVTVGETNRRFVIRADYLNHPVLQELLDQAYEGYGF-NKSGPLSIPCDEFLFE 76
DVP+G+LAV VGE +RFVI YLN QELL Q+ E + + + G L+IPC E +F
Sbjct: 144 DVPKGYLAVYVGEKMKRFVIPVSYLNQTSFQELLSQSEEQFEYDHPMGGLTIPCREDIFL 323
Query: 77 DI 78
DI
Sbjct: 324 DI 329
>TC88867 weakly similar to GP|9758890|dbj|BAB09466.1 auxin-induced
protein-like {Arabidopsis thaliana}, partial (83%)
Length = 453
Score = 58.5 bits (140), Expect = 6e-10
Identities = 31/68 (45%), Positives = 42/68 (61%), Gaps = 2/68 (2%)
Frame = +3
Query: 17 SDVPRGHLAVTVGETNR-RFVIRADYLNHPVLQELLDQAYEGYGFN-KSGPLSIPCDEFL 74
S+VP+GH+AV VGE + RFV+ YLNHP +LL E +G+N G L+IPC E
Sbjct: 138 SNVPKGHIAVYVGELQKKRFVVPISYLNHPTFLDLLSSVEEEFGYNHPMGGLTIPCKEDA 317
Query: 75 FEDILLSL 82
F ++ L
Sbjct: 318 FINLTSQL 341
>BF519656 similar to SP|P32295|ARG7_P Indole-3-acetic acid induced protein
ARG7. [Mung bean Vigna radiata] {Phaseolus aureus},
partial (80%)
Length = 483
Score = 57.8 bits (138), Expect = 1e-09
Identities = 32/68 (47%), Positives = 41/68 (60%), Gaps = 1/68 (1%)
Frame = +2
Query: 18 DVPRGHLAVTVGETNRRFVIRADYLNHPVLQELLDQAYEGYGF-NKSGPLSIPCDEFLFE 76
+VP+G+LAV VGE +RFVI YLN Q LL QA E +G+ + G L+IPC E +F
Sbjct: 131 NVPKGYLAVYVGEQMKRFVIPTSYLNQASFQNLLSQAEEEFGYDHPMGGLTIPCTEDVFL 310
Query: 77 DILLSLGG 84
I G
Sbjct: 311 HITSHFNG 334
>BQ153482 similar to PIR|T06083|T0608 probable auxin-induced protein
T9A14.120 - Arabidopsis thaliana, partial (72%)
Length = 508
Score = 56.6 bits (135), Expect = 2e-09
Identities = 44/117 (37%), Positives = 60/117 (50%), Gaps = 2/117 (1%)
Frame = +3
Query: 7 KNACSGKKLPSDVPRGHLAVTVGETNRRFVIRADYLNHPVLQELLDQAYEGYGF-NKSGP 65
K A S K + VP+G+LAV VGE +RFVI YL QELL Q+ E + + + G
Sbjct: 126 KRASSSKSV--GVPKGYLAVYVGEEMKRFVIPISYLKQKSFQELLSQSEEQFEYDHPMGG 299
Query: 66 LSIPCDEFLFEDILLSLGGGTVARRSSS-PVLTKKLDLSFLKDAVPLLEAFDSKRSN 121
L+IPC E +F DI L P+ L+F + ++ LL D KRS+
Sbjct: 300 LTIPCGEDVFLDITSRLN*RYQKTDIDI*PIFVAL*FLTFKRFSLLLLSYLDVKRSS 470
>TC82431 weakly similar to GP|8777399|dbj|BAA96989.1
emb|CAB86483.1~gene_id:MFB16.16~similar to unknown
protein {Arabidopsis thaliana}, partial (33%)
Length = 717
Score = 56.2 bits (134), Expect = 3e-09
Identities = 26/63 (41%), Positives = 36/63 (56%)
Frame = +1
Query: 20 PRGHLAVTVGETNRRFVIRADYLNHPVLQELLDQAYEGYGFNKSGPLSIPCDEFLFEDIL 79
P G L V VG +RFVI+ NHP+ + LL+ YG+ GPL +PCD LF + L
Sbjct: 265 PHGCLCVYVGPERQRFVIKIKIFNHPLFKTLLEDVENEYGYRNDGPLWLPCDVDLFCEAL 444
Query: 80 LSL 82
+ +
Sbjct: 445 VEI 453
>CB894182 similar to SP|P33083|AX6B_S Auxin-induced protein 6B. [Soybean]
{Glycine max}, partial (88%)
Length = 827
Score = 56.2 bits (134), Expect = 3e-09
Identities = 30/69 (43%), Positives = 43/69 (61%), Gaps = 1/69 (1%)
Frame = -1
Query: 17 SDVPRGHLAVTVGETNRRFVIRADYLNHPVLQELLDQAYEGYGF-NKSGPLSIPCDEFLF 75
++VP+G+LAV VGE +RFVI YLN Q+LL +A + +G+ + G L+IPC E +F
Sbjct: 617 TNVPKGYLAVYVGEEMKRFVIHMSYLNQTSFQDLLSRAEDEFGYDHPMGGLTIPCREEVF 438
Query: 76 EDILLSLGG 84
I G
Sbjct: 437 LHITSRFNG 411
>TC82710 similar to PIR|A84906|A84906 probable auxin-regulated protein
[imported] - Arabidopsis thaliana, partial (58%)
Length = 721
Score = 56.2 bits (134), Expect = 3e-09
Identities = 28/62 (45%), Positives = 39/62 (62%), Gaps = 2/62 (3%)
Frame = +3
Query: 19 VPRGHLAVTVG--ETNRRFVIRADYLNHPVLQELLDQAYEGYGFNKSGPLSIPCDEFLFE 76
VP+G LA+ VG E +RFV+ Y NHP+ +LL +A E YGF+ G ++IPC F
Sbjct: 282 VPKGCLAIKVGQGEDQQRFVVPVIYFNHPLFMQLLKEAEEEYGFDHKGAITIPCRVEEFR 461
Query: 77 DI 78
+I
Sbjct: 462 NI 467
>BQ155897 similar to PIR|T12211|T1221 auxin-induced protein - common ice
plant (fragment), partial (84%)
Length = 632
Score = 56.2 bits (134), Expect = 3e-09
Identities = 30/66 (45%), Positives = 41/66 (61%), Gaps = 10/66 (15%)
Frame = +3
Query: 19 VPRGHLAVTVG--------ETNRRFVIRADYLNHPVLQELLDQAYEGYGFNKSGPLSIPC 70
V +G LAV VG ++FVI YL HP+ + LLDQAY+ YG++ +GPL +PC
Sbjct: 87 VKKGWLAVQVGLEEEEEGGSCAQKFVIPISYLYHPLFKRLLDQAYDVYGYHTNGPLKLPC 266
Query: 71 --DEFL 74
D+FL
Sbjct: 267 SVDDFL 284
>BQ153323 similar to PIR|T06083|T0608 probable auxin-induced protein
T9A14.120 - Arabidopsis thaliana, partial (75%)
Length = 455
Score = 55.8 bits (133), Expect = 4e-09
Identities = 31/68 (45%), Positives = 40/68 (58%), Gaps = 1/68 (1%)
Frame = +1
Query: 18 DVPRGHLAVTVGETNRRFVIRADYLNHPVLQELLDQAYEGYGF-NKSGPLSIPCDEFLFE 76
+VP+G+LAV VGE +RFVI YLN Q LL Q E +G+ + G L+IPC E +F
Sbjct: 58 NVPKGYLAVYVGEQMKRFVIPTSYLNQASFQTLLSQVEEEFGYDHPMGGLTIPCTEDVFL 237
Query: 77 DILLSLGG 84
I G
Sbjct: 238 HITSHFNG 261
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.319 0.138 0.411
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,853,438
Number of Sequences: 36976
Number of extensions: 45359
Number of successful extensions: 209
Number of sequences better than 10.0: 62
Number of HSP's better than 10.0 without gapping: 206
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 206
length of query: 125
length of database: 9,014,727
effective HSP length: 84
effective length of query: 41
effective length of database: 5,908,743
effective search space: 242258463
effective search space used: 242258463
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 52 (24.6 bits)
Medicago: description of AC139525.10


