

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC139355.5 - phase: 0
(672 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
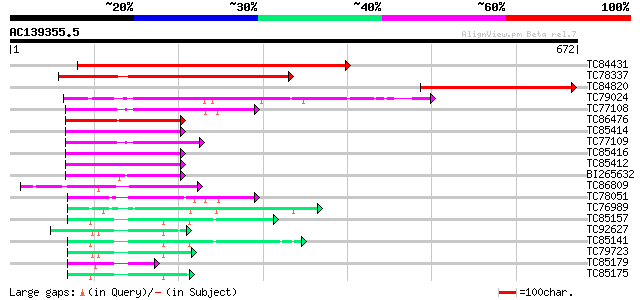
TC84431 similar to GP|10176983|dbj|BAB10215. GTP-binding membran... 636 0.0
TC78337 similar to GP|9759359|dbj|BAB10014.1 GTP-binding protein... 295 5e-80
TC84820 similar to GP|22136082|gb|AAM91119.1 GTP-binding protein... 219 3e-57
TC79024 similar to PIR|A84716|A84716 probable GTP-binding protei... 122 4e-28
TC77108 similar to GP|9758030|dbj|BAB08691.1 GTP-binding protein... 118 8e-27
TC86476 similar to GP|15450763|gb|AAK96653.1 elongation factor E... 114 2e-25
TC85414 homologue to GP|15450763|gb|AAK96653.1 elongation factor... 107 1e-23
TC77109 similar to GP|9758030|dbj|BAB08691.1 GTP-binding protein... 107 1e-23
TC85416 homologue to GP|15450763|gb|AAK96653.1 elongation factor... 106 3e-23
TC85412 homologue to GP|15450763|gb|AAK96653.1 elongation factor... 106 3e-23
BI265632 homologue to GP|22947038|gb CG2238-PA {Drosophila melan... 99 4e-21
TC86809 homologue to PIR|S35701|S35701 translation elongation fa... 86 5e-17
TC78051 homologue to PIR|T01400|T01400 translation elongation fa... 63 3e-10
TC76989 homologue to SP|O24310|EFTU_PEA Elongation factor Tu ch... 61 1e-09
TC85157 homologue to PIR|F86214|F86214 protein T6D22.2 [imported... 61 2e-09
TC92627 similar to PIR|T49975|T49975 hypothetical protein F12B17... 61 2e-09
TC85141 homologue to PIR|F86214|F86214 protein T6D22.2 [imported... 61 2e-09
TC79723 homologue to SP|Q01520|EF1A_PODAN Elongation factor 1-al... 55 9e-08
TC85179 similar to GP|2662343|dbj|BAA23658.1 EF-1 alpha {Oryza s... 52 7e-07
TC85175 homologue to GP|3869088|dbj|BAA34348.1 elongation factor... 52 7e-07
>TC84431 similar to GP|10176983|dbj|BAB10215. GTP-binding membrane protein
LepA homolog {Arabidopsis thaliana}, partial (46%)
Length = 976
Score = 636 bits (1640), Expect = 0.0
Identities = 322/323 (99%), Positives = 323/323 (99%)
Frame = +3
Query: 81 STLADRLLELTGTIKKGLGQPQYLDKLQVERERGITVKAQTATMFYKNIINGDDFKDGKE 140
+TLADRLLELTGTIKKGLGQPQYLDKLQVERERGITVKAQTATMFYKNIINGDDFKDGKE
Sbjct: 3 TTLADRLLELTGTIKKGLGQPQYLDKLQVERERGITVKAQTATMFYKNIINGDDFKDGKE 182
Query: 141 SSNYLLNLIDTPGHVDFSYEVSRSLAACQGVLLVVDAAQGVQAQTVANFYLAFESNLAII 200
SSNYLLNLIDTPGHVDFSYEVSRSLAACQGVLLVVDAAQGVQAQTVANFYLAFESNLAII
Sbjct: 183 SSNYLLNLIDTPGHVDFSYEVSRSLAACQGVLLVVDAAQGVQAQTVANFYLAFESNLAII 362
Query: 201 PVINKIDQPTADPDRVKGQLKSMFDLDPSDALLTSAKTGVGLEHVLPAVIERIPPPPGKS 260
PVINKIDQPTADPDRVKGQLKSMFDLDPSDALLTSAKTGVGLEHVLPAVIERIPPPPGKS
Sbjct: 363 PVINKIDQPTADPDRVKGQLKSMFDLDPSDALLTSAKTGVGLEHVLPAVIERIPPPPGKS 542
Query: 261 ESSLRMLLLDSYFDEYRGVICHVAVVDGALRKGDKISSAATGKSYEAMDIGIMHPELTPT 320
ESSLRMLLLDSYFDEYRGVICHVAVVDGALRKGDKISSAATGKSYEAMDIGIMHPELTPT
Sbjct: 543 ESSLRMLLLDSYFDEYRGVICHVAVVDGALRKGDKISSAATGKSYEAMDIGIMHPELTPT 722
Query: 321 GILFTGQVGYVITGMRTTKEARIGDTIYHTKSTVDVEPLPGFKAAKHMVFSGLFPADGSD 380
GILFTGQVGYVITGMRTTKEARIGDTIYHTKSTVDVEPLPGFKAAKHMVFSGLFPADGSD
Sbjct: 723 GILFTGQVGYVITGMRTTKEARIGDTIYHTKSTVDVEPLPGFKAAKHMVFSGLFPADGSD 902
Query: 381 FEALSHAIEKLTCNDASVSVTKE 403
FEALSHAIEKLTCNDASVSVTKE
Sbjct: 903 FEALSHAIEKLTCNDASVSVTKE 971
>TC78337 similar to GP|9759359|dbj|BAB10014.1 GTP-binding protein LepA
homolog {Arabidopsis thaliana}, partial (40%)
Length = 1035
Score = 295 bits (754), Expect = 5e-80
Identities = 150/278 (53%), Positives = 198/278 (70%)
Frame = +1
Query: 59 LSQYPPELVRNFSIIAHVDHGKSTLADRLLELTGTIKKGLGQPQYLDKLQVERERGITVK 118
LS+ P +RNFSIIAH+DHGKSTLAD+LL++TGT+ + + Q+LD + +ERERGIT+K
Sbjct: 235 LSKVPVSNIRNFSIIAHIDHGKSTLADKLLQVTGTVPQREMKEQFLDNMDLERERGITIK 414
Query: 119 AQTATMFYKNIINGDDFKDGKESSNYLLNLIDTPGHVDFSYEVSRSLAACQGVLLVVDAA 178
Q A M Y E+ Y LNLIDTPGHVDFSYEVSRSLAAC+G LLVVDA+
Sbjct: 415 LQAARMRYVF-----------ENEPYCLNLIDTPGHVDFSYEVSRSLAACEGALLVVDAS 561
Query: 179 QGVQAQTVANFYLAFESNLAIIPVINKIDQPTADPDRVKGQLKSMFDLDPSDALLTSAKT 238
QGV+AQT+AN YLA ++NL IIPV+NKID P A+PDRV +++ + LD S+A+L SAK
Sbjct: 562 QGVEAQTLANVYLALDNNLEIIPVLNKIDLPGAEPDRVLKEIEEVIGLDCSNAILCSAKE 741
Query: 239 GVGLEHVLPAVIERIPPPPGKSESSLRMLLLDSYFDEYRGVICHVAVVDGALRKGDKISS 298
G+G+ +L A++ R+PPP S+ LR L+ DSY+D YRGVI + VVDG ++KGD++
Sbjct: 742 GIGIMDILNAIVARVPPPSDTSKKPLRALIFDSYYDPYRGVIVYFRVVDGTIKKGDRVYF 921
Query: 299 AATGKSYEAMDIGIMHPELTPTGILFTGQVGYVITGMR 336
A+GK Y A +IG++ P LF G VGY+ +R
Sbjct: 922 MASGKDYFADEIGVLSPNQLQAEKLFAG*VGYLSASIR 1035
>TC84820 similar to GP|22136082|gb|AAM91119.1 GTP-binding protein LepA-like
protein {Arabidopsis thaliana}, partial (32%)
Length = 926
Score = 219 bits (557), Expect = 3e-57
Identities = 108/185 (58%), Positives = 142/185 (76%)
Frame = +2
Query: 487 PSEYVGPVITLLSERRGEQLEYSFIDSQRVFMKYRLPLREIVIDFYNELKSITSGYASFD 546
P +Y+GP++ L ERRG+ E F+ R + Y LPL E+V DF+++LKS + GYAS +
Sbjct: 98 PKDYIGPLMELAQERRGQFKEMKFMSEIRASLTYELPLAEMVGDFFDQLKSRSKGYASME 277
Query: 547 YEDSDYQPSDLVKLDILLNGQAVDAMATIVHNSKSYRVGRELVEKLKKVIDRQMFEIIIQ 606
Y + Y+ SDL KLDI +NG+ V+ +ATIVH K+Y VGR L KLK++I RQMF+I IQ
Sbjct: 278 YTVTGYKESDLTKLDIQINGECVEPLATIVHRDKAYSVGRALTLKLKELIPRQMFKIPIQ 457
Query: 607 AAIGSKIIARETVTAMRKNVLAKCYGGDITRKKKLLEKQKEGKKRMKRVGSVDVPQEAFH 666
A IG+K+IA E+++A+RK+VLAKCYGGDITRKKKLL+KQ EGKKRMK +G VDVPQEAF
Sbjct: 458 ACIGAKVIASESLSAIRKDVLAKCYGGDITRKKKLLKKQAEGKKRMKSIGKVDVPQEAFM 637
Query: 667 ELLKV 671
+LK+
Sbjct: 638 AVLKL 652
>TC79024 similar to PIR|A84716|A84716 probable GTP-binding protein
[imported] - Arabidopsis thaliana, partial (67%)
Length = 1513
Score = 122 bits (306), Expect = 4e-28
Identities = 123/475 (25%), Positives = 198/475 (40%), Gaps = 34/475 (7%)
Frame = +2
Query: 64 PELVRNFSIIAHVDHGKSTLADRLLELTGTIKKGLGQPQYLDKLQVERERGITVKAQTAT 123
P +RN ++IAHVDHGK+TL DRLL G + + +D + +ERERGIT+ ++ +
Sbjct: 194 PNRLRNVAVIAHVDHGKTTLMDRLLRQCGA---DIPHERAMDSISLERERGITISSKVTS 364
Query: 124 MFYKNIINGDDFKDGKESSNYLLNLIDTPGHVDFSYEVSRSLAACQGVLLVVDAAQGVQA 183
+ +KD + LN++DTPGH DF EV R + +G +LVVDA +G A
Sbjct: 365 I---------SWKDNE------LNMVDTPGHADFGGEVERVVGMVEGAVLVVDAGEGPLA 499
Query: 184 QTVANFYLAFESNLAIIPVINKIDQPTADPDRVKGQLKSMFDLDPS----------DALL 233
QT A + L I ++NK+D+P + +FDL + L
Sbjct: 500 QTKFVLAKALKYGLRPILLLNKVDRPAVTQEICDEVESVVFDLFANLGATEEQLDFPVLY 679
Query: 234 TSAKTG--------------VGLEHVLPAVIERIPPPPGKSESSLRMLLLDSYFDEYRGV 279
SAK G + +L A++ +PPP ++ +ML+ D Y G
Sbjct: 680 ASAKEGWASTTYTKDPPADAKNMSQLLDAIVSHVPPPNANIDAPFQMLVSMMEKDNYFGR 859
Query: 280 ICHVAVVDGALRKGDKI-----SSAATGKSYEAMDIGIMHPELTPTGILFTGQVGYVITG 334
I V G +R GDK+ + K + + +M + T + G +I+
Sbjct: 860 ILTGRVHSGVVRVGDKVHGLRNKDSGAEKIEDGKVVKVMKKKGTTMVVTDCAGAGDIIS- 1036
Query: 335 MRTTKEARIGDT-----IYHTKSTVDVEPLPGFKAAKHMVFSGLFPADGSDFEALSHAIE 389
+ IG T I TV+++P P + S L DG+
Sbjct: 1037IAGLSSPSIGHTVTTVEIMSALPTVELDP-PTISMTFGVNDSPLAGRDGTHLTGGKIGDR 1213
Query: 390 KLTCNDASVSVTKETSTALGLGFRCGFLGLLHMDVFHQRLEQEYGAHIISTVPTVPYIYE 449
+ ++ ++++ + F G L + + + + +E G + + P V +Y+
Sbjct: 1214LMAESETNLAI--NVLPGMSETFEVQGRGELQLGILIENMRRE-GFELSVSPPKV--MYK 1378
Query: 450 YSDGSKLEVQNPAALPSNPKQRVVACWEPTVIATIVMPSEYVGPVITLLSERRGE 504
G KL EP TI + E+VG V+ LS RR E
Sbjct: 1379TDKGQKL--------------------EPIEEVTIEVNDEHVGFVMEALSHRRAE 1483
>TC77108 similar to GP|9758030|dbj|BAB08691.1 GTP-binding protein typA
(tyrosine phosphorylated protein A) {Arabidopsis
thaliana}, partial (98%)
Length = 2589
Score = 118 bits (295), Expect = 8e-27
Identities = 82/248 (33%), Positives = 128/248 (51%), Gaps = 18/248 (7%)
Frame = +3
Query: 67 VRNFSIIAHVDHGKSTLADRLLELTGTIKKGLG-QPQYLDKLQVERERGITVKAQTATMF 125
+RN +I+AHVDHGK+TL D +L+ T + Q + +D +ERERGIT+ ++ ++
Sbjct: 396 IRNIAIVAHVDHGKTTLVDAMLKQTKVFRDNQTVQERIMDSNDLERERGITILSKNTSVT 575
Query: 126 YKNIINGDDFKDGKESSNYLLNLIDTPGHVDFSYEVSRSLAACQGVLLVVDAAQGVQAQT 185
YK D K +N+IDTPGH DF EV R L +G+LLVVD+ +G QT
Sbjct: 576 YK---------DAK------INIIDTPGHSDFGGEVERILNMVEGILLVVDSVEGPMPQT 710
Query: 186 VANFYLAFESNLAIIPVINKIDQPTADPDRVKGQLKSMF-DLDPSD---------ALLTS 235
A E A++ V+NKID+ +A PD V +F +L+ +D A
Sbjct: 711 RFVLKKALEFGHAVVVVVNKIDRSSARPDFVVNSTFELFIELNATDEQCDFQVIYASGIK 890
Query: 236 AKTGVGLEH-------VLPAVIERIPPPPGKSESSLRMLLLDSYFDEYRGVICHVAVVDG 288
K G+ E + +++ IP P + SL+ML+ + ++ ++G I + G
Sbjct: 891 GKAGLSPEDLADDLGPLFESIMRCIPGPRIDKDGSLQMLVTSTEYEGHKGRIAIGRLEAG 1070
Query: 289 ALRKGDKI 296
L KG ++
Sbjct: 1071VLEKGMEV 1094
Score = 40.4 bits (93), Expect = 0.002
Identities = 22/75 (29%), Positives = 41/75 (54%), Gaps = 1/75 (1%)
Frame = +2
Query: 481 IATIVMPSEYVGPVITLLSERRGEQLEYSFIDSQRV-FMKYRLPLREIVIDFYNELKSIT 539
IAT+ +P E++G V+ LL +RRG+ + + S+ +KY++P R + + N + + +
Sbjct: 1682 IATVEVPEEHMGSVVELLGQRRGQMFDMQGVGSEGTNLLKYKIPTRGL-LGLRNAILTAS 1858
Query: 540 SGYASFDYEDSDYQP 554
G A + Y P
Sbjct: 1859 RGTAILNTIFDSYGP 1903
>TC86476 similar to GP|15450763|gb|AAK96653.1 elongation factor EF-2
{Arabidopsis thaliana}, complete
Length = 2974
Score = 114 bits (284), Expect = 2e-25
Identities = 63/145 (43%), Positives = 91/145 (62%), Gaps = 3/145 (2%)
Frame = +2
Query: 67 VRNFSIIAHVDHGKSTLADRLLELTGTIKKGL-GQPQYLDKLQVERERGITVKAQTATMF 125
+RN S+IAHVDHGKSTL D L+ G I + + G + D Q E ERGIT+K+ +++
Sbjct: 299 IRNMSVIAHVDHGKSTLTDSLVAAAGIIAQEVAGDVRMTDTRQDEAERGITIKSTGISLY 478
Query: 126 YKNIINGD--DFKDGKESSNYLLNLIDTPGHVDFSYEVSRSLAACQGVLLVVDAAQGVQA 183
Y+ + +GD +FK +E + YL+NLID+PGHVDFS EV+ +L G L+VVD +GV
Sbjct: 479 YE-MSDGDLKNFKGEREGNKYLINLIDSPGHVDFSSEVTAALRITDGALVVVDCVEGVCV 655
Query: 184 QTVANFYLAFESNLAIIPVINKIDQ 208
QT A + + +NK+D+
Sbjct: 656 QTETVLRQALGERIKPVLTVNKMDR 730
>TC85414 homologue to GP|15450763|gb|AAK96653.1 elongation factor EF-2
{Arabidopsis thaliana}, complete
Length = 2892
Score = 107 bits (267), Expect = 1e-23
Identities = 59/144 (40%), Positives = 85/144 (58%), Gaps = 2/144 (1%)
Frame = +3
Query: 67 VRNFSIIAHVDHGKSTLADRLLELTGTIKKGL-GQPQYLDKLQVERERGITVKAQTATMF 125
+RN S+IAHVDHGKSTL D L+ G I + + G + D E ERGIT+K+ +++
Sbjct: 180 IRNMSVIAHVDHGKSTLTDSLVAAAGIIAQEVAGDVRMTDTRADEAERGITIKSTGISLY 359
Query: 126 YKNIING-DDFKDGKESSNYLLNLIDTPGHVDFSYEVSRSLAACQGVLLVVDAAQGVQAQ 184
Y+ + FK + + YL+NLID+PGHVDFS EV+ +L G L+VVD +GV Q
Sbjct: 360 YEMTDDSLKSFKGERNGNEYLINLIDSPGHVDFSSEVTAALRITDGALVVVDCVEGVCVQ 539
Query: 185 TVANFYLAFESNLAIIPVINKIDQ 208
T A + + +NK+D+
Sbjct: 540 TETVLRQALGERIRPVLTVNKMDR 611
>TC77109 similar to GP|9758030|dbj|BAB08691.1 GTP-binding protein typA
(tyrosine phosphorylated protein A) {Arabidopsis
thaliana}, partial (27%)
Length = 672
Score = 107 bits (267), Expect = 1e-23
Identities = 65/166 (39%), Positives = 95/166 (57%), Gaps = 2/166 (1%)
Frame = +3
Query: 67 VRNFSIIAHVDHGKSTLADRLLELTGTIKKGLG-QPQYLDKLQVERERGITVKAQTATMF 125
+RN +I+AHVDHGK+TL D +L+ T + Q + +D +ERERGIT+ ++ ++
Sbjct: 186 IRNIAIVAHVDHGKTTLVDAMLKQTKVFRDNQTVQERIMDSNDLERERGITILSKNTSVT 365
Query: 126 YKNIINGDDFKDGKESSNYLLNLIDTPGHVDFSYEVSRSLAACQGVLLVVDAAQGVQAQT 185
YK D K +N+IDTPGH DF EV R L +G+LLVVD+ +G QT
Sbjct: 366 YK---------DAK------INIIDTPGHSDFGGEVERILNMVEGILLVVDSVEGPMPQT 500
Query: 186 VANFYLAFESNLAIIPVINKIDQPTADPDRVKGQLKSMF-DLDPSD 230
A E A++ V+NKID+ +A PD V +F +L+ +D
Sbjct: 501 RFVLKKALEFGHAVVVVVNKIDRSSARPDFVVNSTFELFIELNATD 638
>TC85416 homologue to GP|15450763|gb|AAK96653.1 elongation factor EF-2
{Arabidopsis thaliana}, partial (21%)
Length = 660
Score = 106 bits (265), Expect = 3e-23
Identities = 59/144 (40%), Positives = 84/144 (57%), Gaps = 2/144 (1%)
Frame = +3
Query: 67 VRNFSIIAHVDHGKSTLADRLLELTGTIKKGL-GQPQYLDKLQVERERGITVKAQTATMF 125
+RN S+IAHVDHGKSTL D L+ G I + + G + D E ERGIT+K+ +++
Sbjct: 165 IRNMSVIAHVDHGKSTLTDSLVAAAGIIAQEVAGDVRMTDTRADEAERGITIKSTGISLY 344
Query: 126 YKNIING-DDFKDGKESSNYLLNLIDTPGHVDFSYEVSRSLAACQGVLLVVDAAQGVQAQ 184
Y+ FK + + YL+NLID+PGHVDFS EV+ +L G L+VVD +GV Q
Sbjct: 345 YEMSDESLKSFKGERNGNEYLINLIDSPGHVDFSSEVTAALRITDGALVVVDCVEGVCVQ 524
Query: 185 TVANFYLAFESNLAIIPVINKIDQ 208
T A + + +NK+D+
Sbjct: 525 TETVLRQALGERIRPVLTVNKMDR 596
>TC85412 homologue to GP|15450763|gb|AAK96653.1 elongation factor EF-2
{Arabidopsis thaliana}, complete
Length = 3229
Score = 106 bits (264), Expect = 3e-23
Identities = 59/144 (40%), Positives = 84/144 (57%), Gaps = 2/144 (1%)
Frame = +3
Query: 67 VRNFSIIAHVDHGKSTLADRLLELTGTIKKGL-GQPQYLDKLQVERERGITVKAQTATMF 125
+RN S+IAHVDHGKSTL D L+ G I + + G + D E ERGIT+K+ +++
Sbjct: 222 IRNMSVIAHVDHGKSTLTDSLVAAAGIIAQEVAGDVRMTDTRADEAERGITIKSTGISLY 401
Query: 126 YKNIING-DDFKDGKESSNYLLNLIDTPGHVDFSYEVSRSLAACQGVLLVVDAAQGVQAQ 184
Y+ FK + + YL+NLID+PGHVDFS EV+ +L G L+VVD +GV Q
Sbjct: 402 YEMTDESLKRFKGERNGNEYLINLIDSPGHVDFSSEVTAALRITDGALVVVDCVEGVCVQ 581
Query: 185 TVANFYLAFESNLAIIPVINKIDQ 208
T A + + +NK+D+
Sbjct: 582 TETVLRQALGERIRPVLTVNKMDR 653
>BI265632 homologue to GP|22947038|gb CG2238-PA {Drosophila melanogaster},
partial (21%)
Length = 593
Score = 99.4 bits (246), Expect = 4e-21
Identities = 58/149 (38%), Positives = 84/149 (55%), Gaps = 7/149 (4%)
Frame = +3
Query: 67 VRNFSIIAHVDHGKSTLADRLLELTGTIKKG-LGQPQYLDKLQVERERGITVKAQTATMF 125
+RN S+IAHVDHGKSTL D L+ G I G+ ++ D + E++R IT+K+ +MF
Sbjct: 111 IRNMSVIAHVDHGKSTLTDSLVSKAGIIAGARAGETRFTDTRKDEQDRCITIKSTAISMF 290
Query: 126 YKN------IINGDDFKDGKESSNYLLNLIDTPGHVDFSYEVSRSLAACQGVLLVVDAAQ 179
++ I D ++ K +L+NLID+PGHVDFS EV+ +L G L+VVD
Sbjct: 291 FELEEKDLVFITNPDQRE-KSEKGFLINLIDSPGHVDFSSEVTAALRVTDGALVVVDCVS 467
Query: 180 GVQAQTVANFYLAFESNLAIIPVINKIDQ 208
GV QT A + +NK+D+
Sbjct: 468 GVCVQTETVLRQAIAERIKPXLFMNKMDR 554
>TC86809 homologue to PIR|S35701|S35701 translation elongation factor EF-G
chloroplast - soybean, partial (91%)
Length = 2674
Score = 85.9 bits (211), Expect = 5e-17
Identities = 63/219 (28%), Positives = 104/219 (46%), Gaps = 4/219 (1%)
Frame = +2
Query: 14 SNYVSLLFNFNPLSSRITHERFSITRALFCTQSRQNYTKEKAIIDLSQYPPELVRNFSII 73
S+ +S F +P+ T + TR F + + K + L Y RN I+
Sbjct: 179 SSSLSQFFRTSPIKP--TSPQLVRTRRNFSVFAMSTPDEAKRAVPLKDY-----RNIGIM 337
Query: 74 AHVDHGKSTLADRLLELTGTIKKGLGQPQY----LDKLQVERERGITVKAQTATMFYKNI 129
AH+D GK+T +R+L TG K +G+ +D ++ E+ERGIT+ + T F+
Sbjct: 338 AHIDAGKTTTTERILFYTGRNYK-IGEVHEGTATMDWMEQEQERGITITSAATTTFW--- 505
Query: 130 INGDDFKDGKESSNYLLNLIDTPGHVDFSYEVSRSLAACQGVLLVVDAAQGVQAQTVANF 189
N+ +N+IDTPGHVDF+ EV R+L G + + D+ GV+ Q+ +
Sbjct: 506 ------------DNHRINIIDTPGHVDFTLEVERALRVLDGAICLFDSVAGVEPQSETVW 649
Query: 190 YLAFESNLAIIPVINKIDQPTADPDRVKGQLKSMFDLDP 228
A + I +NK+D+ A+ R + + + P
Sbjct: 650 RQADRYGVPRICFVNKMDRLGANFFRTRDMIVTNLGAKP 766
Score = 37.4 bits (85), Expect = 0.019
Identities = 51/237 (21%), Positives = 90/237 (37%), Gaps = 25/237 (10%)
Frame = +2
Query: 235 SAKTGVGLEHVLPAVIERIPPP----------PGKSESSLRMLLLDS----------YFD 274
SA G++ +L AV++ +P P P E+++ + D D
Sbjct: 1070 SAFKNKGVQPLLDAVVDYLPSPLDVPPMKGTDPENPEATIERIAGDDEPFSGLAFKIMSD 1249
Query: 275 EYRGVICHVAVVDGALRKGDKISSAATGKSYEAMDIGIMHPELTP-TGILFTGQVGYVIT 333
+ G + V V G L G + ++ GK + MH + TG I
Sbjct: 1250 SFVGSLTFVRVYSGKLTAGSYVLNSNKGKKERIGRLLEMHANSREDVKVALTGD----IV 1417
Query: 334 GMRTTKEARIGDTIYHTKSTVDVE----PLPGFKAAKHMVFSGLFPADGSDFEALSHAIE 389
+ K+ G+T+ +S V +E P P K A + P +D + ++ +
Sbjct: 1418 ALAGLKDTITGETLCDPESPVVLERMDFPDPVIKIA-------IEPKTKADIDKMAAGLV 1576
Query: 390 KLTCNDASVSVTKETSTALGLGFRCGFLGLLHMDVFHQRLEQEYGAHIISTVPTVPY 446
KL D S +++ + +G LH+++ RL++EY P V Y
Sbjct: 1577 KLAQEDPSFHFSRDEEINQTV---IEGMGELHLEIIVDRLKREYKVEANVGAPQVNY 1738
>TC78051 homologue to PIR|T01400|T01400 translation elongation factor EF-Tu
precursor mitochondrial - Arabidopsis thaliana, partial
(87%)
Length = 1681
Score = 63.2 bits (152), Expect = 3e-10
Identities = 64/248 (25%), Positives = 104/248 (41%), Gaps = 20/248 (8%)
Frame = +3
Query: 69 NFSIIAHVDHGKSTLADRLLELTGTIKKGLGQP-QYLDKLQVERERGITVKAQTATMFYK 127
N I HVDHGK+TL + + K +DK E++RGIT+ TA + Y
Sbjct: 237 NVGTIGHVDHGKTTLTAAITRVLADEGKAKAVAFDEIDKAPEEKKRGITIA--TAHVEY- 407
Query: 128 NIINGDDFKDGKESSNYLLNLIDTPGHVDFSYEVSRSLAACQGVLLVVDAAQGVQAQTVA 187
E++ +D PGH D+ + A G +LVV A G QT
Sbjct: 408 ------------ETAKRHYAHVDCPGHADYVKNMITGAAQMDGGILVVSAPDGPMPQTKE 551
Query: 188 NFYLAFESNL-AIIPVINKIDQPTADPDRVK------GQLKSMFDLDPSD---------A 231
+ LA + + +++ +NK+D DP+ ++ +L S + D +
Sbjct: 552 HILLARQVGVPSLVCFLNKVD-AVDDPELLELVEMELRELLSFYKFPGDDIPIIRGSALS 728
Query: 232 LLTSAKTGVGLEHVL---PAVIERIPPPPGKSESSLRMLLLDSYFDEYRGVICHVAVVDG 288
L +G + +L AV E IP P + E M + D + + RG + V G
Sbjct: 729 ALQGTNEEIGRKAILKLMDAVDEYIPDPVRQLEKPFLMPIEDVFSIQGRGTVATGRVEQG 908
Query: 289 ALRKGDKI 296
++ GD++
Sbjct: 909 IIKIGDEV 932
>TC76989 homologue to SP|O24310|EFTU_PEA Elongation factor Tu chloroplast
precursor (EF-Tu). [Garden pea] {Pisum sativum}, partial
(92%)
Length = 1907
Score = 61.2 bits (147), Expect = 1e-09
Identities = 76/337 (22%), Positives = 129/337 (37%), Gaps = 35/337 (10%)
Frame = +3
Query: 69 NFSIIAHVDHGKSTLADRLLELTGTIKKGLGQPQYLDKLQV---ERERGITVKAQTATMF 125
N I HVDHGK+TL L ++ G P+ D++ ER RGIT+ TAT+
Sbjct: 423 NIGTIGHVDHGKTTLTAALTMALASL--GNSAPKKYDEIDAAPEERARGITIN--TATVE 590
Query: 126 YKNIINGDDFKDGKESSNYLLNLIDTPGHVDFSYEVSRSLAACQGVLLVVDAAQGVQAQT 185
Y+ E+ +Y +D PGH D+ + A G +LVV A G QT
Sbjct: 591 YET-----------ETRHYAH--VDCPGHADYVKNMITGAAQMDGAILVVSGADGPMPQT 731
Query: 186 VANFYLAFESNL-AIIPVINKIDQPTADP-----DRVKGQLKSMFDLDPSDALLTSAKTG 239
+ LA + + +++ +NK DQ + + +L S ++ D + S
Sbjct: 732 KEHILLAKQVGVPSVVVFLNKQDQVDDEELLELVELEVRELLSSYEFPGDDIPIVSGSAL 911
Query: 240 VGLE--------------------HVLPAVIERIPPPPGKSESSLRMLLLDSYFDEYRGV 279
+ LE ++ V IP P ++E + + D + RG
Sbjct: 912 LALEALVANPTIKRGDNEWVDKIYKLMDEVDNYIPIPQRQTELPFLLAIEDVFSITGRGT 1091
Query: 280 ICHVAVVDGALRKGDKISSAATGKSYEAMDIGIMHPELTPTGILFTGQVGYVITGM---- 335
+ + G ++ GD + ++ G+ + + VG ++ G+
Sbjct: 1092VATGRIERGLVKVGDVVDLVGLRETRSTTVTGVEMFQKILDDAMAGDNVGLLLRGVQKID 1271
Query: 336 --RTTKEARIGDTIYHTKSTVDVEPLPGFKAAKHMVF 370
R A+ G H+K V L + +H F
Sbjct: 1272IQRGMVLAKPGTITPHSKFECIVYVLKKEEGGRHSPF 1382
>TC85157 homologue to PIR|F86214|F86214 protein T6D22.2 [imported] -
Arabidopsis thaliana, partial (46%)
Length = 1738
Score = 60.8 bits (146), Expect = 2e-09
Identities = 74/297 (24%), Positives = 109/297 (35%), Gaps = 47/297 (15%)
Frame = +1
Query: 69 NFSIIAHVDHGKSTLADRLLELTGTI----------------KKGLGQPQYLDKLQVERE 112
N +I HVD GKST L+ G I K+ LDKL+ ERE
Sbjct: 106 NIVVIGHVDSGKSTTTGHLIYKLGGIDKRVIERFEKEAAEMNKRSFKYAWVLDKLKAERE 285
Query: 113 RGITVKAQTATMFYKNIINGDDFKDGKESSNYLLNLIDTPGHVDFSYEVSRSLAACQGVL 172
RGIT+ E++ Y +ID PGH DF + + +
Sbjct: 286 RGITIDIALWKF---------------ETTKYYCTVIDAPGHRDFIKNMITGTSQADCAV 420
Query: 173 LVVDAAQG-------VQAQTVANFYLAFESNL-AIIPVINKIDQPTA------------- 211
L++D+ G QT + LAF + +I NK+D T
Sbjct: 421 LIIDSTTGGFEAGISKDGQTREHALLAFTLGVKQMICCCNKMDATTPKYSKGRYDEIVKE 600
Query: 212 ----------DPDRVKGQLKSMFDLDPSDALLTSAKTGVGLEHVLPAVIERIPPPPGKSE 261
+PD++ S F+ D T+ G L +++I P S+
Sbjct: 601 VSSYLKKVGYNPDKIPFVPISGFEGDNMIERSTNLDWYKG--PTLLEALDQINEPKRPSD 774
Query: 262 SSLRMLLLDSYFDEYRGVICHVAVVDGALRKGDKISSAATGKSYEAMDIGIMHPELT 318
LR+ L D Y G + V G ++ G I+ A TG + E + + H LT
Sbjct: 775 KPLRLPLQDVYKIGGIGTVPVGRVETGVIKPGMVITFAPTGLTTEVKSVEMHHEALT 945
>TC92627 similar to PIR|T49975|T49975 hypothetical protein F12B17.10 -
Arabidopsis thaliana, partial (25%)
Length = 827
Score = 60.8 bits (146), Expect = 2e-09
Identities = 54/193 (27%), Positives = 79/193 (39%), Gaps = 26/193 (13%)
Frame = +2
Query: 49 NYTKEKAIIDLSQYPPELVRNFSIIAHVDHGKSTLADRLLELTGTIKK------------ 96
+Y EK ++ N +I+ HVD GKSTL+ RLL L G I K
Sbjct: 164 SYQPEKWMLPQQSEDTLTQLNLAIVGHVDSGKSTLSGRLLHLLGRISKKEMHKYEKEAKL 343
Query: 97 -GLGQPQY---LDKLQVERERGITVKAQTATMFYKNIINGDDFKDGKESSNYLLNLIDTP 152
G G Y LD+ ERERGIT+ A ++ Y + ++D+P
Sbjct: 344 QGKGSFAYAWALDESSEERERGITMTVAVAYF---------------DTKKYHVVVLDSP 478
Query: 153 GHVDFSYEVSRSLAACQGVLLVVDAAQGV----------QAQTVANFYLAFESNLAIIPV 202
GH DF + +LV+DA+ G Q + A +F + I+
Sbjct: 479 GHKDFIPNMISGATQADAAVLVIDASLGAFEAGMDGGKGQTREHAQLIRSFGVDHVIV-A 655
Query: 203 INKIDQPTADPDR 215
+NK+D + DR
Sbjct: 656 VNKMDAVSYSKDR 694
>TC85141 homologue to PIR|F86214|F86214 protein T6D22.2 [imported] -
Arabidopsis thaliana, partial (47%)
Length = 1881
Score = 60.8 bits (146), Expect = 2e-09
Identities = 80/330 (24%), Positives = 122/330 (36%), Gaps = 47/330 (14%)
Frame = +3
Query: 69 NFSIIAHVDHGKSTLADRLLELTGTI----------------KKGLGQPQYLDKLQVERE 112
N +I HVD GKST L+ G I K+ LDKL+ ERE
Sbjct: 102 NIVVIGHVDSGKSTTTGHLIYKLGGIDKRVIERFEKEAAEMNKRSFKYAWVLDKLKAERE 281
Query: 113 RGITVKAQTATMFYKNIINGDDFKDGKESSNYLLNLIDTPGHVDFSYEVSRSLAACQGVL 172
RGIT+ E++ Y +ID PGH DF + + +
Sbjct: 282 RGITIDIALWKF---------------ETTKYYCTVIDAPGHRDFIKNMITGTSQADCAV 416
Query: 173 LVVDAAQG-------VQAQTVANFYLAFESNL-AIIPVINKIDQPTA------------- 211
L++D+ G QT + LAF + +I NK+D T
Sbjct: 417 LIIDSTTGGFEAGISKDGQTREHALLAFTLGVRQMICCCNKMDATTPKYSKSRYDEIVKE 596
Query: 212 ----------DPDRVKGQLKSMFDLDPSDALLTSAKTGVGLEHVLPAVIERIPPPPGKSE 261
+PD++ S F+ D T+ G L +++I P S+
Sbjct: 597 VSSYMKKVGYNPDKIPFVPISGFEGDNMIERSTNLDWYKG--PTLLEALDQINEPKRPSD 770
Query: 262 SSLRMLLLDSYFDEYRGVICHVAVVDGALRKGDKISSAATGKSYEAMDIGIMHPELTPTG 321
LR+ L D Y G + V G ++ G ++ A TG E + + H LT
Sbjct: 771 KPLRLPLQDVYKIGGIGTVPVGRVETGTMKPGMVVTFAPTGLQTEVKSVEMHHESLTEA- 947
Query: 322 ILFTGQVGYVITGMRTTKEARIGDTIYHTK 351
L VG+ + + + K+ + G +TK
Sbjct: 948 -LPGDNVGFNVKNV-SVKDLKRGYVASNTK 1031
>TC79723 homologue to SP|Q01520|EF1A_PODAN Elongation factor 1-alpha
(EF-1-alpha). {Podospora anserina}, complete
Length = 1537
Score = 55.1 bits (131), Expect = 9e-08
Identities = 51/177 (28%), Positives = 71/177 (39%), Gaps = 24/177 (13%)
Frame = +1
Query: 69 NFSIIAHVDHGKSTLADRLLELTGTIKK-------------GLGQPQY---LDKLQVERE 112
N +I HVD GKST L+ G I K G G +Y LDKL+ ERE
Sbjct: 103 NVVVIGHVDSGKSTTTGHLIYQCGGIDKRTIEKFEKEAAELGKGSFKYAWVLDKLKAERE 282
Query: 113 RGITVKAQTATMFYKNIINGDDFKDGKESSNYLLNLIDTPGHVDFSYEVSRSLAACQGVL 172
RGIT+ E+ Y + +ID PGH DF + + +
Sbjct: 283 RGITIDIALWKF---------------ETPKYYVTVIDAPGHRDFIKNMITGTSQADCAV 417
Query: 173 LVVDAAQG-------VQAQTVANFYLAFESNL-AIIPVINKIDQPTADPDRVKGQLK 221
L++ A G QT + LA+ + +I INK+D DR + +K
Sbjct: 418 LIIAAGTGEFEAGISKDGQTREHALLAYTLGVKQLIVAINKMDTTKWSEDRYQEIIK 588
>TC85179 similar to GP|2662343|dbj|BAA23658.1 EF-1 alpha {Oryza sativa},
partial (96%)
Length = 1789
Score = 52.0 bits (123), Expect = 7e-07
Identities = 39/125 (31%), Positives = 51/125 (40%), Gaps = 16/125 (12%)
Frame = +2
Query: 69 NFSIIAHVDHGKSTLADRLLELTGTIKKGLGQ----------------PQYLDKLQVERE 112
N I HVD GKST A L+ G IKK + + LDKL+ ERE
Sbjct: 176 NIVFIGHVDSGKSTTAGHLIYKLGGIKKDVIERFEKEAAEINMCSFKYAWVLDKLKAERE 355
Query: 113 RGITVKAQTATMFYKNIINGDDFKDGKESSNYLLNLIDTPGHVDFSYEVSRSLAACQGVL 172
RGIT+ + E++ Y LID PGH DF + + +
Sbjct: 356 RGITIDISLSKF---------------ETTKYYCTLIDAPGHRDFIKNMITGTSQADCAV 490
Query: 173 LVVDA 177
LV D+
Sbjct: 491 LVYDS 505
>TC85175 homologue to GP|3869088|dbj|BAA34348.1 elongation factor-1 alpha
{Nicotiana paniculata}, partial (53%)
Length = 831
Score = 52.0 bits (123), Expect = 7e-07
Identities = 49/175 (28%), Positives = 68/175 (38%), Gaps = 24/175 (13%)
Frame = +3
Query: 69 NFSIIAHVDHGKSTLADRLLELTGTI----------------KKGLGQPQYLDKLQVERE 112
N +I HVD GKST L+ G I K+ LDKL+ ERE
Sbjct: 132 NIVVIGHVDSGKSTTTGHLIYKLGGIDKRVIERFEKEAAEMNKRSFKYAWVLDKLKAERE 311
Query: 113 RGITVKAQTATMFYKNIINGDDFKDGKESSNYLLNLIDTPGHVDFSYEVSRSLAACQGVL 172
RGIT+ E++ Y +ID PGH DF + + +
Sbjct: 312 RGITIDIALWKF---------------ETTKYYCTVIDAPGHRDFIKNMITGTSQADCAV 446
Query: 173 LVVDAAQG-------VQAQTVANFYLAFESNL-AIIPVINKIDQPTADPDRVKGQ 219
L++D+ G QT + LAF + +I NK+D T P KG+
Sbjct: 447 LIIDSTTGGFEAGISKDGQTREHALLAFTLGVKQMICCCNKMDATT--PKYSKGR 605
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.319 0.136 0.383
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 17,204,818
Number of Sequences: 36976
Number of extensions: 215163
Number of successful extensions: 1084
Number of sequences better than 10.0: 64
Number of HSP's better than 10.0 without gapping: 1040
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1051
length of query: 672
length of database: 9,014,727
effective HSP length: 103
effective length of query: 569
effective length of database: 5,206,199
effective search space: 2962327231
effective search space used: 2962327231
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 62 (28.5 bits)
Medicago: description of AC139355.5


