

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC139343.2 + phase: 0 /pseudo
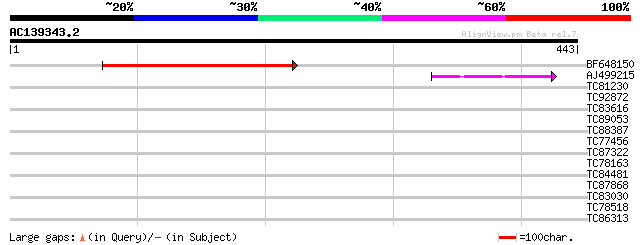
(443 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BF648150 similar to GP|14586969|gb| pol polyprotein {Citrus x pa... 116 2e-26
AJ499215 weakly similar to GP|6642775|gb| gag-pol polyprotein {V... 47 1e-05
TC81230 40 0.001
TC92872 38 0.007
TC83616 similar to GP|16226661|gb|AAL16226.1 At2g28910/F8N16.20 ... 32 0.64
TC89053 similar to GP|17104693|gb|AAL34235.1 unknown protein {Ar... 30 1.9
TC88387 similar to GP|13357253|gb|AAK20050.1 putative zinc finge... 30 1.9
TC77456 similar to PIR|T10580|T10580 zinc-finger protein Lsd1 - ... 30 2.4
TC87322 similar to GP|12322716|gb|AAG51340.1 unknown protein; 28... 30 2.4
TC78163 H+-ATPase 28 5.4
TC84481 similar to GP|11875626|gb|AAG40731.1 XRN4 {Arabidopsis t... 28 7.1
TC87868 similar to PIR|T05112|T05112 splicing factor 9G8-like SR... 28 7.1
TC83030 weakly similar to GP|18855061|gb|AAL79753.1 putative RNA... 28 7.1
TC78518 similar to PIR|JC7234|JC7234 27k vesicle-associated memb... 28 7.1
TC86313 similar to GP|4995890|emb|CAB44316.1 serine palmitoyltra... 24 7.1
>BF648150 similar to GP|14586969|gb| pol polyprotein {Citrus x paradisi},
partial (3%)
Length = 658
Score = 116 bits (291), Expect = 2e-26
Identities = 50/153 (32%), Positives = 96/153 (62%)
Frame = +2
Query: 73 KPDEAKSDAKQIEKWNNANKVCRHTILSTISDALFDVYCSYKVAKDIWDNMNVKYTAEEA 132
K DE ++ + +KW+N + +C IL+ +SD+LFD+Y S AKD+WD + +Y E+A
Sbjct: 137 KDDETVAETRDRQKWDNDDYICLGHILNGMSDSLFDIYQSSPSAKDLWDKLETRYMREDA 316
Query: 133 TKQKFVVGNYL*WQMKEDKEIKVQINEYHQLIEELKAENIILPDVFVAAALVEKLRSSWD 192
T +KF+V ++ ++M ++K + Q+ E +++ K N+ + + + +++++KL SW
Sbjct: 317 TSKKFLVSHFNNYKMVDNKSVMEQLYEIERILNNYKQHNMNMDETIIVSSIIDKLPPSWK 496
Query: 193 DYKQQLKHKHKQMSLNDLIRHIIIEDTSRKECG 225
D+K+ +KHK + +SL L H+ + + RK+ G
Sbjct: 497 DFKRTMKHKKEDISLEQLGNHLRLXEEYRKQEG 595
>AJ499215 weakly similar to GP|6642775|gb| gag-pol polyprotein {Vitis
vinifera}, partial (18%)
Length = 567
Score = 47.0 bits (110), Expect = 1e-05
Identities = 32/98 (32%), Positives = 48/98 (48%)
Frame = +3
Query: 330 WVVDSGATRHICAKREAFTSYISVGDDEDQVYLGDSRTTAVNGKGKVILKLTSGKTLALS 389
W++DSG T H+ R+ F +V + + V G G V++K SG +S
Sbjct: 108 WLIDSGCTHHMTHDRDLFKELNK--STISKVRMLNGAHIEVEGIGTVLVKSHSGYK-QIS 278
Query: 390 DVLNVPTIRTNLISVALLNKVGVKVSFESDKMVMSKNN 427
+VL P + +L+SV L G KV FE +K V+ N
Sbjct: 279 NVLYAPKLNQSLLSVPQLLTKGYKVLFEHEKCVIKDQN 392
>TC81230
Length = 958
Score = 40.4 bits (93), Expect = 0.001
Identities = 51/250 (20%), Positives = 94/250 (37%), Gaps = 8/250 (3%)
Frame = +1
Query: 43 VFKGQNFRRWNERVFTLLD-------VHGVASVLTDVKPDEAKSDAKQIEKWNNANKVCR 95
+ G N+ W E + L V G T K D A + A ++E+W++ N
Sbjct: 235 ILNGSNYNHWAESMCGFLKGRRLWRYVTGDKKCPTKGKDDTADAFADKLEEWDSKNHQII 414
Query: 96 HTILSTISDALFDVYCSYKVAKDIWDNMNVKYTAEEATKQKFVVGNYL*WQMKEDKEIKV 155
+T ++ + ++ AK++WD++ +YT + + Q + +K+ +K
Sbjct: 415 TWFRNTSIPSIHMQFGRFENAKEVWDHLKQRYTISDLSHQYQL--------LKDLSNLKQ 570
Query: 156 QINEYHQLIEELKAENIILPDVFVAAALVEKLRSSWDDYKQQLKHKHKQMSLNDLIRHI- 214
Q Q + E A+ ++ + + E D K H+++ + L+
Sbjct: 571 QSG---QPVYEFLAQMEVIWNQLTSC---EPSLKDATDMKTYETHRNRVRLIQFLMALTD 732
Query: 215 IIEDTSRKECGAARAKALESRANLIQNNARKQQRYENKSDHALKVTNPNFKANLGECYVC 274
E LE+ +++ + Q K+D A VTN K C C
Sbjct: 733 EYEPVRASSLHQNPLPTLENALPCLKSEETRLQLVPPKADLAFAVTNNATK----PCRHC 900
Query: 275 GKKGHKAYHC 284
K GH C
Sbjct: 901 QKSGHSFSDC 930
>TC92872
Length = 923
Score = 38.1 bits (87), Expect = 0.007
Identities = 38/169 (22%), Positives = 71/169 (41%), Gaps = 8/169 (4%)
Frame = +1
Query: 16 DSTFSVATNPPAVVTYAKQFPDVSRIEVFKGQNFRRWNERVFTLL-------DVHGVASV 68
D F++ATNP T + + S +NFR W +V TLL + G +
Sbjct: 31 DPIFNMATNPSN--TKSSLILNASITFKLSRKNFRAWKRQVTTLLAGIEVMGHIDGTTPI 204
Query: 69 LTD-VKPDEAKSDAKQIEKWNNANKVCRHTILSTISDALFDVYCSYKVAKDIWDNMNVKY 127
T+ + + + KW +++ + +LS++++A + SY+ A+ +W + ++
Sbjct: 205 QTETIINNGVSAPNPDYTKWFTLDQLIINLLLSSMTEADSISFASYETARTLWVAIESQF 384
Query: 128 TAEEATKQKFVVGNYL*WQMKEDKEIKVQINEYHQLIEELKAENIILPD 176
+ V N + K DK I + L +EL + L D
Sbjct: 385 N-NTSRSHVMSVTNQIQRCTKGDKSITDYLFSVKSLADELAVIDKSLSD 528
>TC83616 similar to GP|16226661|gb|AAL16226.1 At2g28910/F8N16.20
{Arabidopsis thaliana}, partial (30%)
Length = 854
Score = 31.6 bits (70), Expect = 0.64
Identities = 17/60 (28%), Positives = 26/60 (43%)
Frame = +2
Query: 258 KVTNPNFKANLGECYVCGKKGHKAYHCRYRKVNGQPPKPKANLTQGDDKKYDDDDVIAAV 317
K+TN + G C CG+ GH + C K N+ DDK+ D D + ++
Sbjct: 362 KITNSEVDVSRGACKKCGRVGHLKFQC------------KNNVKIKDDKEEKDFDAMQSL 505
>TC89053 similar to GP|17104693|gb|AAL34235.1 unknown protein {Arabidopsis
thaliana}, partial (57%)
Length = 1327
Score = 30.0 bits (66), Expect = 1.9
Identities = 12/21 (57%), Positives = 15/21 (71%)
Frame = +3
Query: 41 IEVFKGQNFRRWNERVFTLLD 61
+ V+KGQ F W E+VF LLD
Sbjct: 270 VNVYKGQRFPVWKEQVFHLLD 332
>TC88387 similar to GP|13357253|gb|AAK20050.1 putative zinc finger protein
{Oryza sativa (japonica cultivar-group)}, partial (96%)
Length = 1286
Score = 30.0 bits (66), Expect = 1.9
Identities = 14/34 (41%), Positives = 17/34 (49%)
Frame = +1
Query: 267 NLGECYVCGKKGHKAYHCRYRKVNGQPPKPKANL 300
N G C+ CGK GH+A C P KP +L
Sbjct: 583 NEGICHTCGKAGHRARECTV------PQKPPGDL 666
>TC77456 similar to PIR|T10580|T10580 zinc-finger protein Lsd1 - Arabidopsis
thaliana, partial (90%)
Length = 1472
Score = 29.6 bits (65), Expect = 2.4
Identities = 18/59 (30%), Positives = 27/59 (45%)
Frame = +2
Query: 335 GATRHICAKREAFTSYISVGDDEDQVYLGDSRTTAVNGKGKVILKLTSGKTLALSDVLN 393
GAT CA T+ G D Q+Y G RT + +G ++ + T+ L+ V N
Sbjct: 353 GATNVCCALCNTITAVPPPGMDMSQLYCGGCRTLLMYTRGATSVRCSCCHTVNLAPVSN 529
>TC87322 similar to GP|12322716|gb|AAG51340.1 unknown protein; 28504-31237
{Arabidopsis thaliana}, partial (62%)
Length = 1356
Score = 29.6 bits (65), Expect = 2.4
Identities = 14/52 (26%), Positives = 27/52 (51%)
Frame = -3
Query: 65 VASVLTDVKPDEAKSDAKQIEKWNNANKVCRHTILSTISDALFDVYCSYKVA 116
V + T +K E + + + K+N +N +TIL + D+Y +YK++
Sbjct: 1315 VKLIFTHIK--EIHTPSTNLSKYNKSNNCVNYTILLVQLNVTIDIYAAYKLS 1166
>TC78163 H+-ATPase
Length = 3295
Score = 28.5 bits (62), Expect = 5.4
Identities = 21/72 (29%), Positives = 35/72 (48%), Gaps = 3/72 (4%)
Frame = +2
Query: 133 TKQKFVVGNYL*WQMKEDKEIKVQ-INEYH--QLIEELKAENIILPDVFVAAALVEKLRS 189
TK + V + WQ + E++++ +H QLI+EL ++LP + A V + R
Sbjct: 1265 TKMQLTVQLFQCWQTQRRHELELKKFISFHSIQLIKELHLHTLMLPVICTGLAKVHQSRF 1444
Query: 190 SWDDYKQQLKHK 201
S +Q HK
Sbjct: 1445 SILHVTKQKLHK 1480
>TC84481 similar to GP|11875626|gb|AAG40731.1 XRN4 {Arabidopsis thaliana},
partial (23%)
Length = 727
Score = 28.1 bits (61), Expect = 7.1
Identities = 10/22 (45%), Positives = 13/22 (58%)
Frame = +2
Query: 264 FKANLGECYVCGKKGHKAYHCR 285
F +CY+CG+ GH A CR
Sbjct: 188 FPGQQEKCYMCGQVGHLAAECR 253
>TC87868 similar to PIR|T05112|T05112 splicing factor 9G8-like SR protein
RSZp22 [validated] - Arabidopsis thaliana, partial (91%)
Length = 860
Score = 28.1 bits (61), Expect = 7.1
Identities = 11/22 (50%), Positives = 13/22 (59%)
Frame = +3
Query: 270 ECYVCGKKGHKAYHCRYRKVNG 291
+CY CG+ GH A CR R G
Sbjct: 333 KCYECGEPGHFARECRNRGGGG 398
>TC83030 weakly similar to GP|18855061|gb|AAL79753.1 putative RNA helicase
{Oryza sativa}, partial (7%)
Length = 624
Score = 28.1 bits (61), Expect = 7.1
Identities = 10/25 (40%), Positives = 17/25 (68%)
Frame = +1
Query: 260 TNPNFKANLGECYVCGKKGHKAYHC 284
++PN ++ G C+ CG+ GH+A C
Sbjct: 136 SSPN-RSFAGTCFTCGESGHRASDC 207
>TC78518 similar to PIR|JC7234|JC7234 27k vesicle-associated membrane
protein-associated protein - curled-leaved tobacco,
partial (83%)
Length = 827
Score = 28.1 bits (61), Expect = 7.1
Identities = 12/36 (33%), Positives = 17/36 (46%)
Frame = +1
Query: 278 GHKAYHCRYRKVNGQPPKPKANLTQGDDKKYDDDDV 313
GH C+ R V PP P + + +G D+ D V
Sbjct: 418 GHNVEECKLRVVYVAPPGPPSPVREGSDEDSSPDSV 525
>TC86313 similar to GP|4995890|emb|CAB44316.1 serine palmitoyltransferase
{Solanum tuberosum}, partial (98%)
Length = 2052
Score = 24.3 bits (51), Expect(2) = 7.1
Identities = 9/21 (42%), Positives = 13/21 (61%)
Frame = +3
Query: 40 RIEVFKGQNFRRWNERVFTLL 60
+I F GQN RRW+ R ++
Sbjct: 1341 KIVTFSGQNCRRWDLRFLEIM 1403
Score = 21.9 bits (45), Expect(2) = 7.1
Identities = 12/44 (27%), Positives = 22/44 (49%)
Frame = +2
Query: 93 VCRHTILSTISDALFDVYCSYKVAKDIWDNMNVKYTAEEATKQK 136
+ R I + S + D+ + +V + D + +KY E TKQ+
Sbjct: 1523 LARARICISASHSREDLTKALQVISSVGDLIGIKYFPAEPTKQQ 1654
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.317 0.132 0.384
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,278,336
Number of Sequences: 36976
Number of extensions: 150102
Number of successful extensions: 805
Number of sequences better than 10.0: 30
Number of HSP's better than 10.0 without gapping: 796
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 805
length of query: 443
length of database: 9,014,727
effective HSP length: 99
effective length of query: 344
effective length of database: 5,354,103
effective search space: 1841811432
effective search space used: 1841811432
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 60 (27.7 bits)
Medicago: description of AC139343.2


