

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC138580.8 - phase: 0
(268 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
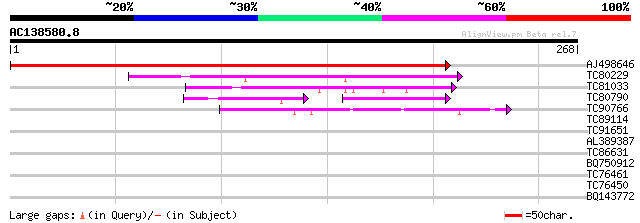
Score E
Sequences producing significant alignments: (bits) Value
AJ498646 similar to PIR|E96759|E96 probable tumor-related protei... 410 e-115
TC80229 similar to PIR|H86215|H86215 protein T6D22.22 [imported]... 114 4e-26
TC81033 similar to PIR|T02654|T02654 hypothetical protein At2g26... 64 8e-11
TC80790 similar to PIR|T38541|T38541 probable sucrose carrier - ... 33 2e-05
TC90766 similar to PIR|T05491|T05491 hypothetical protein T19K4.... 44 9e-05
TC89114 similar to GP|14517476|gb|AAK62628.1 At2g43320/T1O24.6 {... 39 0.003
TC91651 SP|P40389|UV22_SCHPO UV-induced protein uvi22. [Fission ... 38 0.004
AL389387 similar to GP|9759518|dbj| gene_id:MLN1.9~pir||T38261~s... 37 0.010
TC86631 similar to GP|7407189|gb|AAF61950.1| phosphoethanolamine... 29 2.2
BQ750912 similar to GP|7291124|gb| CG15311 gene product {Drosoph... 28 3.7
TC76461 homologue to PIR|T09570|T09570 carbonate dehydratase (EC... 27 6.3
TC76450 similar to GP|17065130|gb|AAL32719.1 putative protein {A... 27 6.3
BQ143772 similar to SP|O04979|LON1_ Lon protease homolog 1 mito... 27 8.3
>AJ498646 similar to PIR|E96759|E96 probable tumor-related protein T9L24.46
[imported] - Arabidopsis thaliana, partial (50%)
Length = 665
Score = 410 bits (1054), Expect = e-115
Identities = 208/208 (100%), Positives = 208/208 (100%)
Frame = +2
Query: 1 MEEEEENATAAMVKLGSYGGSVMLVVPGEESAAEETMLLWGIQQPTLSKPNAFVAQSSLQ 60
MEEEEENATAAMVKLGSYGGSVMLVVPGEESAAEETMLLWGIQQPTLSKPNAFVAQSSLQ
Sbjct: 41 MEEEEENATAAMVKLGSYGGSVMLVVPGEESAAEETMLLWGIQQPTLSKPNAFVAQSSLQ 220
Query: 61 LRLDSCGHSLSILQSPSSLGKPGVTGSVMWDSGIVLGKFLEHSVDSGMLVLQGKKIVELG 120
LRLDSCGHSLSILQSPSSLGKPGVTGSVMWDSGIVLGKFLEHSVDSGMLVLQGKKIVELG
Sbjct: 221 LRLDSCGHSLSILQSPSSLGKPGVTGSVMWDSGIVLGKFLEHSVDSGMLVLQGKKIVELG 400
Query: 121 SGCGLVGCIAALLGGEVILTDLPDRMRLLRKNIETNMKHISLRGSITATELTWGDDPDQE 180
SGCGLVGCIAALLGGEVILTDLPDRMRLLRKNIETNMKHISLRGSITATELTWGDDPDQE
Sbjct: 401 SGCGLVGCIAALLGGEVILTDLPDRMRLLRKNIETNMKHISLRGSITATELTWGDDPDQE 580
Query: 181 LIDPTPDYILGSDVVYSEGAVVDLLETL 208
LIDPTPDYILGSDVVYSEGAVVDLLETL
Sbjct: 581 LIDPTPDYILGSDVVYSEGAVVDLLETL 664
>TC80229 similar to PIR|H86215|H86215 protein T6D22.22 [imported] -
Arabidopsis thaliana, partial (19%)
Length = 697
Score = 114 bits (285), Expect = 4e-26
Identities = 63/166 (37%), Positives = 100/166 (59%), Gaps = 8/166 (4%)
Frame = +3
Query: 57 SSLQLRLDSCGHSLSILQSPSSLGKPGVTGSVMWDSGIVLGKFLEHSVDSGMLV---LQG 113
++ ++ L+ GH L Q P+S G+ +WD+ +V KFLE + G L+G
Sbjct: 153 TTFEMPLEVLGHDLLFAQDPNSKHH----GTTIWDASLVFAKFLERNCRKGRFSPAKLKG 320
Query: 114 KKIVELGSGCGLVGCIAALLGGEVILTDLPDRMRLLRKNIETNM-----KHISLRGSITA 168
K+++ELG+GCG+ G A+LG +VI+TD + + LL++N++ N+ K+ L GSI
Sbjct: 321 KRVIELGAGCGVSGFAMAMLGCDVIVTDQKEVLPLLQRNVDRNISRVMQKNPELFGSIKV 500
Query: 169 TELTWGDDPDQELIDPTPDYILGSDVVYSEGAVVDLLETLGQLSGP 214
+EL WGD+ + + P DYI+G+DVVY E + LL+ + LSGP
Sbjct: 501 SELQWGDESHIKAVGPPFDYIIGTDVVYVEHLLEPLLQKILALSGP 638
>TC81033 similar to PIR|T02654|T02654 hypothetical protein At2g26810
[imported] - Arabidopsis thaliana, partial (54%)
Length = 697
Score = 63.5 bits (153), Expect = 8e-11
Identities = 39/136 (28%), Positives = 76/136 (55%), Gaps = 8/136 (5%)
Frame = +3
Query: 84 VTGSVMWDSGIVLGKFLEHSVDSGMLVLQGKKIVELGSGCGLVGCIAALLGGEVILTDLP 143
+TG ++W ++L +L ++ + QG +ELGSG G+ G + +V+LTD
Sbjct: 183 LTGQLVWPGAMLLNDYLSKHIE----MFQGCTAIELGSGVGITGILCRRFCNKVVLTDHN 350
Query: 144 DR-MRLLRKNIETNM--KHIS-LRGSITATELTWGD-DPDQELIDPTP---DYILGSDVV 195
+ +++++KNIE + ++IS + A +L WG+ D E++ P D++LG+D+
Sbjct: 351 EEVLKIIKKNIELHSCPENISPTSNGLVAEKLEWGNTDQIHEILQKHPGGFDFVLGADIC 530
Query: 196 YSEGAVVDLLETLGQL 211
+ + + L +T+ QL
Sbjct: 531 FQQSNIPLLFDTVRQL 578
>TC80790 similar to PIR|T38541|T38541 probable sucrose carrier - fission
yeast (Schizosaccharomyces pombe), partial (3%)
Length = 1461
Score = 33.1 bits (74), Expect(2) = 2e-05
Identities = 22/60 (36%), Positives = 31/60 (51%), Gaps = 1/60 (1%)
Frame = +3
Query: 83 GVTGSVMWDSGIVLGKFLEHSVDSGMLVLQGKKIVELGSGCGLVG-CIAALLGGEVILTD 141
G TG +W S + FL + S + K E+GSG GLVG C+A + + IL+D
Sbjct: 510 GDTGCSIWPSSL----FLSELILSHPELFSNKVCFEIGSGVGLVGLCLAHVKASKGILSD 677
Score = 32.0 bits (71), Expect(2) = 2e-05
Identities = 14/51 (27%), Positives = 25/51 (48%)
Frame = +1
Query: 158 KHISLRGSITATELTWGDDPDQELIDPTPDYILGSDVVYSEGAVVDLLETL 208
K + ++ + L W + +L D PD ILG+DV+Y + L+ +
Sbjct: 754 KEMKIQAPVKCMYLPWESASESQLQDIIPDVILGADVIYDPVCLPHLVRVI 906
>TC90766 similar to PIR|T05491|T05491 hypothetical protein T19K4.120 -
Arabidopsis thaliana, partial (63%)
Length = 1320
Score = 43.5 bits (101), Expect = 9e-05
Identities = 41/144 (28%), Positives = 67/144 (46%), Gaps = 6/144 (4%)
Frame = +1
Query: 100 LEHSVDSGMLVLQGKKIVELGSGCGLVGCIAALL--GGEVILTD-LPDRMRLLRKNIETN 156
L H S + + KK++ELGSG GL G + A + EV+++D P + ++NIE N
Sbjct: 379 LAHYCLSHRDIFRSKKVIELGSGYGLAGFVIAAITEASEVVISDGNPQVVDYTQRNIEAN 558
Query: 157 MKHISLRGSITATELTWGDDPDQELIDPTPDYILGSDVVYSEGAVVDLLETLGQL---SG 213
+ + +L W + + D D I+ SD + + DL + L +
Sbjct: 559 SGAFG-DTVVKSMKLHWNQEDTSSVADAF-DIIVASDCTFFKDFHRDLARIVKHLLSKTE 732
Query: 214 PNTTIFLAGELRNDAILEYFLEAA 237
+ IFL+ + N L+ FLE A
Sbjct: 733 SSEAIFLSPKRGNS--LDLFLEVA 798
>TC89114 similar to GP|14517476|gb|AAK62628.1 At2g43320/T1O24.6 {Arabidopsis
thaliana}, partial (58%)
Length = 941
Score = 38.5 bits (88), Expect = 0.003
Identities = 17/49 (34%), Positives = 26/49 (52%)
Frame = +2
Query: 90 WDSGIVLGKFLEHSVDSGMLVLQGKKIVELGSGCGLVGCIAALLGGEVI 138
W S I L L++ + G L +GK+++EL GL G A L G ++
Sbjct: 392 WSSSIDLVSVLKNEIRDGQLTFRGKRVLELSCNYGLPGIFACLKGASIV 538
>TC91651 SP|P40389|UV22_SCHPO UV-induced protein uvi22. [Fission yeast]
{Schizosaccharomyces pombe}, partial (3%)
Length = 655
Score = 38.1 bits (87), Expect = 0.004
Identities = 32/115 (27%), Positives = 54/115 (46%), Gaps = 2/115 (1%)
Frame = +2
Query: 63 LDSCGHSLSI-LQSPSSLGKPGVTGSVMWDSGIVLGKFLEHSVDSGMLVLQGKKIVELGS 121
+ SC +S + +Q + P V G +W + +VL F+ H G +ELG+
Sbjct: 227 ISSCKNSYCVRIQHNITSSIPNV-GLQVWRAELVLTDFILHKALCSS-EFHGVIALELGA 400
Query: 122 GCGLVGCIAALLGGEVILTDLPDR-MRLLRKNIETNMKHISLRGSITATELTWGD 175
G GLVG + A V +TD ++ + KN++ N ++ +I +L W D
Sbjct: 401 GTGLVGLLLARTANSVFVTDRGNQILDNCVKNVQLNRGLLNNPATIFVRDLDWFD 565
>AL389387 similar to GP|9759518|dbj| gene_id:MLN1.9~pir||T38261~similar to
unknown protein {Arabidopsis thaliana}, partial (28%)
Length = 394
Score = 36.6 bits (83), Expect = 0.010
Identities = 21/68 (30%), Positives = 38/68 (55%), Gaps = 1/68 (1%)
Frame = +3
Query: 123 CGLVGCIAALLG-GEVILTDLPDRMRLLRKNIETNMKHISLRGSITATELTWGDDPDQEL 181
CG+ G LLG ++ILTD+P M L+KN+++N L+ ++ + L W +
Sbjct: 3 CGVAGMGLYLLGLTDIILTDIPPVMPALKKNLKSNKP--VLKKNLKYSILYWNNKDQINA 176
Query: 182 IDPTPDYI 189
++P D++
Sbjct: 177 VNPPFDFV 200
>TC86631 similar to GP|7407189|gb|AAF61950.1| phosphoethanolamine
N-methyltransferase {Spinacia oleracea}, partial (39%)
Length = 692
Score = 28.9 bits (63), Expect = 2.2
Identities = 15/52 (28%), Positives = 28/52 (53%)
Frame = +3
Query: 112 QGKKIVELGSGCGLVGCIAALLGGEVILTDLPDRMRLLRKNIETNMKHISLR 163
+GK ++ELG+G G A G+++ D + ++KN TN H +++
Sbjct: 243 EGKSVLELGAGIGRFTAELAQKAGQLLAVDFIE--SAIKKNENTNGHHKNVK 392
>BQ750912 similar to GP|7291124|gb| CG15311 gene product {Drosophila
melanogaster}, partial (2%)
Length = 682
Score = 28.1 bits (61), Expect = 3.7
Identities = 20/50 (40%), Positives = 25/50 (50%), Gaps = 2/50 (4%)
Frame = -2
Query: 40 WGIQQPTLSKPNA--FVAQSSLQLRLDSCGHSLSILQSPSSLGKPGVTGS 87
WG QQ + PNA + +Q S S SL SPS + +P VTGS
Sbjct: 519 WGQQQLAMGSPNASSYGSQPSPS-GSGSSRDSLPATHSPSLMHEPSVTGS 373
>TC76461 homologue to PIR|T09570|T09570 carbonate dehydratase (EC 4.2.1.1) -
alfalfa, complete
Length = 1504
Score = 27.3 bits (59), Expect = 6.3
Identities = 17/51 (33%), Positives = 26/51 (50%), Gaps = 2/51 (3%)
Frame = +1
Query: 210 QLSGPNTTIFLAGELRNDAILEYFLEAAMNDFTI--GRVDQTLWHPDYHSN 258
Q GP TIF EL D ++++ A+ + GR+ + WH +HSN
Sbjct: 274 QKPGPLQTIF---ELHPDEVVDHSYSCALERSFLYHGRMYVSTWHICFHSN 417
>TC76450 similar to GP|17065130|gb|AAL32719.1 putative protein {Arabidopsis
thaliana}, partial (83%)
Length = 2171
Score = 27.3 bits (59), Expect = 6.3
Identities = 17/51 (33%), Positives = 26/51 (50%), Gaps = 2/51 (3%)
Frame = +1
Query: 210 QLSGPNTTIFLAGELRNDAILEYFLEAAMNDFTI--GRVDQTLWHPDYHSN 258
Q GP TIF EL D ++++ A+ + GR+ + WH +HSN
Sbjct: 913 QKPGPLQTIF---ELHPDEVVDHSYSCALERSFLYHGRMYVSTWHICFHSN 1056
>BQ143772 similar to SP|O04979|LON1_ Lon protease homolog 1 mitochondrial
precursor (EC 3.4.21.-). [Spinach] {Spinacia oleracea},
partial (8%)
Length = 682
Score = 26.9 bits (58), Expect = 8.3
Identities = 12/33 (36%), Positives = 18/33 (54%)
Frame = -2
Query: 22 VMLVVPGEESAAEETMLLWGIQQPTLSKPNAFV 54
V++ VPG E ++ + L WG+QQ P V
Sbjct: 642 VLISVPGTEISSVDVALNWGVQQYRNKSPARIV 544
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.316 0.136 0.393
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,362,626
Number of Sequences: 36976
Number of extensions: 91716
Number of successful extensions: 401
Number of sequences better than 10.0: 26
Number of HSP's better than 10.0 without gapping: 399
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 399
length of query: 268
length of database: 9,014,727
effective HSP length: 94
effective length of query: 174
effective length of database: 5,538,983
effective search space: 963783042
effective search space used: 963783042
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 57 (26.6 bits)
Medicago: description of AC138580.8


