

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC138579.1 - phase: 0
(881 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
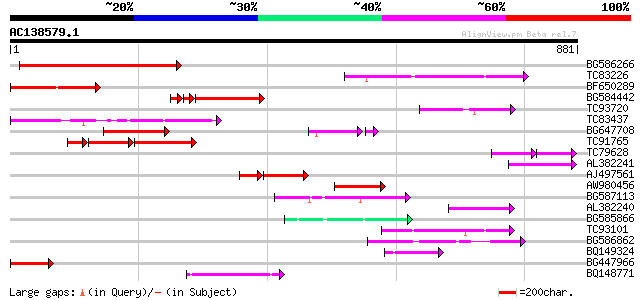
Score E
Sequences producing significant alignments: (bits) Value
BG586266 similar to GP|7267666|em RNA-directed DNA polymerase-li... 218 6e-57
TC83226 weakly similar to PIR|G86419|G86419 probable reverse tra... 120 2e-27
BF650289 weakly similar to GP|9049283|dbj| orf129a {Beta vulgari... 113 3e-25
BG584442 105 5e-25
TC93720 similar to GP|21743320|dbj|BAC03314. putative nematode r... 106 3e-23
TC83437 weakly similar to PIR|D86384|D86384 unknown protein [imp... 104 2e-22
BG647708 weakly similar to GP|13786450|gb| putative reverse tran... 102 8e-22
TC91765 weakly similar to GP|19881779|gb|AAM01180.1 Putative ret... 80 3e-21
TC79628 similar to GP|17104799|gb|AAL34288.1 putative 2-nitropro... 65 1e-18
AL382241 91 3e-18
AJ497561 weakly similar to GP|10140689|gb putative non-LTR retro... 74 6e-18
AW980456 89 1e-17
BG587113 weakly similar to PIR|A84888|A8 hypothetical protein At... 84 3e-16
AL382240 77 2e-14
BG585866 77 2e-14
TC93101 75 8e-14
BG586862 74 2e-13
BQ149324 72 9e-13
BG447966 weakly similar to PIR|G96509|G96 protein F27F5.21 [impo... 71 2e-12
BQ148771 69 1e-11
>BG586266 similar to GP|7267666|em RNA-directed DNA polymerase-like protein
{Arabidopsis thaliana}, partial (18%)
Length = 789
Score = 218 bits (556), Expect = 6e-57
Identities = 111/252 (44%), Positives = 159/252 (63%)
Frame = -3
Query: 16 MKDWRPIALCNVVYKIAAKVLANRLKRVLDKCISDNQSPFVPGRSILDNAMAAIELVHYK 75
+ ++R IA CN YKI AK+L+ R++ +L IS +QS FVPGR+I DN + +++HY
Sbjct: 778 VSEYRTIAPCNTQYKIIAKILSKRMQPLLRSIISPSQSAFVPGRAISDNVLITHKILHYL 599
Query: 76 KTKVKGTQGDVAFKLDISKAYDRIDWNYLKCVMLKMGFSIQWVKWIMMCVETVDYSVIVN 135
+ +A K D++KAYDRI WN+L+ V+ ++GF W+ WIM CV TV YS ++N
Sbjct: 598 RQSGAKKHVSMAVKTDMTKAYDRIAWNFLREVLTRLGFHGIWISWIMECVSTVSYSFLIN 419
Query: 136 EEIAGPIIHGRGLRQGDPLSTYLFIICAEGLSSLIRKAEASGTIHGVKICNNAPIVSHLL 195
G ++ RGLRQGDPLS YLFI+C E LS L ++A GT+ GVK+ N P ++HLL
Sbjct: 418 GGPQGRVLPSRGLRQGDPLSPYLFILCTEVLSGLCQQALRKGTLPGVKVARNCPPINHLL 239
Query: 196 FADDCFLFFRAEPSEAMVMKNILEVNESASGQAINYQKSEIFFSRNGSQSKQLNIPNILQ 255
FADD F ++ S ++ +I++ +ASG+ IN KS I FS SQ+ + L+
Sbjct: 238 FADDTMFFGKSNASSCAILLSIMDKYRAASGRCIN*TKSAITFSSKTSQAIIDRVKGELK 59
Query: 256 VQAVLGTGKYLG 267
+ GTGKYLG
Sbjct: 58 IAKEGGTGKYLG 23
>TC83226 weakly similar to PIR|G86419|G86419 probable reverse transcriptase
100033-105622 [imported] - Arabidopsis thaliana, partial
(2%)
Length = 885
Score = 120 bits (301), Expect = 2e-27
Identities = 85/296 (28%), Positives = 136/296 (45%), Gaps = 10/296 (3%)
Frame = +3
Query: 520 VWNKENNGEYSVRSAYR-LCMQKLLDISEFKVLG----AWDYIWNLKVPPKVKNLVWRVA 574
+W G YSV+S Y L + I+ W IW+L P+ K L+WR+
Sbjct: 3 MWMHNPTGIYSVKSGYNTLRTWQTQQINNTSTSSDETLIWKKIWSLHTIPRHKVLLWRIL 182
Query: 575 RNILPTRMRLREKIVNCPPHCVLCNSADEDSLHLFFNCPSSSNIWNLSNLSNDVKIVVSQ 634
+ LP R LR++ + C P C C+S E HLF +CP S +W SNL +
Sbjct: 183 NDSLPVRSSLRKRGIQCYPLCPRCHSKTETITHLFMSCPLSKRVWFGSNL-----CINFD 347
Query: 635 GLNSSAVIFHLLQVLSAGESAL---FATILWSIWKQRNNKIWNDVTDAQNFVLERASSLL 691
L + I L + + + + A I++++W RN + D T + +++RAS+ +
Sbjct: 348 NLPNPNFIN*LYEAIL*KDECITI*IAAIIYNLWHARNLSVLEDQTILEMDIIQRASNCI 527
Query: 692 YDWNAARNARLNNNVVNGDGEHSQ--STRVIKWSKPNRGRMKCNIDASFPNNENRIGIGI 749
D+ A + G SQ + KW +PN G +K N DA+ N + G+GI
Sbjct: 528 SDYKQANTQAPPSMARTGYDPRSQHRPAKNTKWKRPNLGLVKVNTDANL-QNHGKWGLGI 704
Query: 750 CIRDDEGAFVIARTKCLTPKCDVHIGEALGLLTALQWVHELQLGPIEFELDSKKVV 805
IRD+ G + A T EA LLT +++ + + FE D++K++
Sbjct: 705 IIRDEVGLVMAASTWETDGNDRALEAEAYALLTGMRFAKDCGFXKVXFEGDNEKLM 872
>BF650289 weakly similar to GP|9049283|dbj| orf129a {Beta vulgaris}, partial
(69%)
Length = 616
Score = 113 bits (283), Expect = 3e-25
Identities = 56/140 (40%), Positives = 88/140 (62%)
Frame = +3
Query: 2 TNIALIPKGDMQVSMKDWRPIALCNVVYKIAAKVLANRLKRVLDKCISDNQSPFVPGRSI 61
T + L+PK S+K++RPIA C+V+YKI +K+L +R++ VL+ +S+NQS FV GR I
Sbjct: 201 TYVTLLPKEVNVTSVKNFRPIACCSVIYKIISKILTSRMQGVLNSVVSENQSAFVKGRVI 380
Query: 62 LDNAMAAIELVHYKKTKVKGTQGDVAFKLDISKAYDRIDWNYLKCVMLKMGFSIQWVKWI 121
DN + + ELV K KG K+D+ KAYD +W ++K +ML++GF ++V W+
Sbjct: 381 FDNIILSHELV--KSYSRKGISPRCMVKIDLXKAYDSXEWPFIKHLMLELGFPYKFVNWV 554
Query: 122 MMCVETVDYSVIVNEEIAGP 141
M + T Y+ N ++ P
Sbjct: 555 MAXLTTASYTFNXNGDLTXP 614
>BG584442
Length = 775
Score = 105 bits (261), Expect(3) = 5e-25
Identities = 55/108 (50%), Positives = 69/108 (62%)
Frame = +1
Query: 289 KKINSWSSKCLSKAGREVLMKSMLQSIPTYFMSIFTLPTSLCDEIEKMLNSFWWGHSGTQ 348
KKIN +KCLSK EV++K LQSI +Y MSIF L S DEIEK++N+F W H G
Sbjct: 379 KKINF*RNKCLSKVM*EVMIKYALQSISSYVMSIFLLLNSQVDEIEKIMNTFSWVHVGEN 558
Query: 349 GRGLHWLSWDRLAAHKNDGGMSFKSLPVFNLAMLGKQGWRSMTNPDTL 396
+G+HW+S ++L HKN GGM F FN+ MLGKQ + N TL
Sbjct: 559 RKGMHWMS*EKLFVHKNYGGMGFTDFTTFNIPMLGKQV*SFLLNRTTL 702
Score = 24.6 bits (52), Expect(3) = 5e-25
Identities = 12/19 (63%), Positives = 13/19 (68%)
Frame = +2
Query: 250 IPNILQVQAVLGTGKYLGL 268
I IL V+AVLG KY GL
Sbjct: 239 ITYILDVRAVLGRDKYFGL 295
Score = 23.9 bits (50), Expect(3) = 5e-25
Identities = 10/17 (58%), Positives = 12/17 (69%)
Frame = +3
Query: 271 MTGRSKKATFNFIKDRI 287
M S KATF++IKD I
Sbjct: 324 MLSTSNKATFSYIKDEI 374
>TC93720 similar to GP|21743320|dbj|BAC03314. putative nematode
resistance-like protein {Oryza sativa (japonica
cultivar-group)}, partial (3%)
Length = 457
Score = 106 bits (265), Expect = 3e-23
Identities = 58/158 (36%), Positives = 84/158 (52%), Gaps = 9/158 (5%)
Frame = -2
Query: 637 NSSAVIFHLLQVLSAGESALFATILWSIWKQRNNKIWNDVTDAQNFVLERASSLLYDWNA 696
N +A+IF LLQ S+ + L AT++WS+WK K+W ++ + V+ A+ LL DW A
Sbjct: 453 NINALIFTLLQQFSSDQCELMATVMWSLWKSCRMKLWQQNNESNSQVVHHATHLLNDWRA 274
Query: 697 ARNARLNNNVVNGDGEHSQSTRVIK---------WSKPNRGRMKCNIDASFPNNENRIGI 747
A+ +N H QS V W KCNIDASF + N++G+
Sbjct: 273 AQTI*SHN--------HDQSNMVYPCPMKQDEEAWMTLTPDMYKCNIDASFFTSLNKVGL 118
Query: 748 GICIRDDEGAFVIARTKCLTPKCDVHIGEALGLLTALQ 785
G+C+ DD FV+A+T P CD+ GEA+ L T L+
Sbjct: 117 GMCLADDADDFVLAKTIWFAPSCDIDAGEAVRLHTTLE 4
>TC83437 weakly similar to PIR|D86384|D86384 unknown protein [imported] -
Arabidopsis thaliana, partial (6%)
Length = 951
Score = 104 bits (259), Expect = 2e-22
Identities = 99/339 (29%), Positives = 152/339 (44%), Gaps = 12/339 (3%)
Frame = +2
Query: 2 TNIALIPKGDMQVSMKDWRPIALCNVVYKIAAKVLANRLKRVLDKCISDNQSPFVPGRSI 61
T IALIPK D + D+RPI+L +YKI K+LANRL+ V+ ISD QS FV R I
Sbjct: 59 TFIALIPKVDNPQRLNDFRPISLVGSLYKILGKLLANRLRVVIGSVISDAQSAFVKNRQI 238
Query: 62 LDNAMAAIELVHYKKTKVKGTQGDVAFKLDISKAYDRIDWNYLKCVMLKMG--------- 112
L+ + + + K + + W + + L +G
Sbjct: 239 LEMVFL*QMRLWMRLRN*R-------------KIFCCLRWILKRLITLSIGLIWILF*VG 379
Query: 113 --FSIQWVKWIMMCVETVDYSVIVNEEIAGPIIHGRGLRQGDPLSTYLFIICAEGLSSLI 170
F + W KWI CV T SV+VN G P + + SL+
Sbjct: 380 MSFLVLWRKWIKECVSTATTSVLVN---------------GSPTNVLM--------KSLV 490
Query: 171 RKAEASGTIHGVKICNNAPIVSHLLFADDCFLFFRAEPSEAMVMKNILEVNESASGQAIN 230
+ + GV N +VSHL FA+D L + ++ L + + SG +N
Sbjct: 491 QTQLFTRYSFGVV---NPVVVSHLQFANDTLLLETKNWANIRALRAALVIF*AMSGLKVN 661
Query: 231 YQKSEIFFSRNGSQSKQLNIPNILQVQAVLGTGKYLGLPSMTGRSKKATF-NFIKDRIWK 289
+ KS + N + S ++L + YLG+P + G S++ +F I +RI
Sbjct: 662 FHKSGLVCV-NIAPSWLSEAASVLSWKVGKVPFLYLGMP-IEGNSRRLSFWEPIVNRIKA 835
Query: 290 KINSWSSKCLSKAGREVLMKSMLQSIPTYFMSIFTLPTS 328
++ W+S+ LS GR VL+KS+L S +S++ LP+S
Sbjct: 836 RLTGWNSRFLSFGGRLVLLKSVLTS-----LSVYALPSS 937
>BG647708 weakly similar to GP|13786450|gb| putative reverse transcriptase
{Oryza sativa}, partial (9%)
Length = 708
Score = 102 bits (253), Expect = 8e-22
Identities = 48/104 (46%), Positives = 73/104 (70%), Gaps = 1/104 (0%)
Frame = +1
Query: 146 RGLRQGDPLSTYLFIICAEGLSSLIRKAEASGTIHGVKICNNAPIVSHLLFADDCFLFFR 205
+GLRQGDPLS YLFI+CA LS L+++ +HG+++ + P ++HLLFADD LF R
Sbjct: 10 KGLRQGDPLSPYLFILCANVLSGLLKREGNKQNLHGIQVARSDPKITHLLFADDSLLFAR 189
Query: 206 AEPSEAMVMKNILEVNESASGQAINYQKSEIFFSRN-GSQSKQL 248
A +EA + +L +SASGQ +N++KSE+ +S+N +Q K++
Sbjct: 190 ANLTEAATIMQVLHSYQSASGQLVNFEKSEVSYSQNVPNQEKEM 321
Score = 40.0 bits (92), Expect(2) = 7e-05
Identities = 27/88 (30%), Positives = 42/88 (47%), Gaps = 5/88 (5%)
Frame = +3
Query: 465 PTVGDFHLSH----LRVSHCMLQHHKMWDVPFLETIFDQQTVAAIIKTSLYTSIKEDRFV 520
P+ +F L+H L V + K W+ + F+ IIK L ED+ +
Sbjct: 354 PSKREFKLNHVSGSLSVDELIDYDTKQWNRDLIFHSFNNYAAHQIIKIPLSMRQPEDKII 533
Query: 521 WNKENNGEYSVRSAYR-LCMQKLLDISE 547
W+ E +G YSVRSA+ LC + + +E
Sbjct: 534 WHWEKDGIYSVRSAHHALCDENFSNQAE 617
Score = 25.0 bits (53), Expect(2) = 7e-05
Identities = 8/20 (40%), Positives = 11/20 (55%)
Frame = +2
Query: 554 WDYIWNLKVPPKVKNLVWRV 573
W IWN V+N +WR+
Sbjct: 644 WKAIWNTPASRSVQNFLWRL 703
>TC91765 weakly similar to GP|19881779|gb|AAM01180.1 Putative retroelement
{Oryza sativa (japonica cultivar-group)}, partial (1%)
Length = 625
Score = 79.7 bits (195), Expect = 4e-15
Identities = 46/97 (47%), Positives = 59/97 (60%)
Frame = +3
Query: 194 LLFADDCFLFFRAEPSEAMVMKNILEVNESASGQAINYQKSEIFFSRNGSQSKQLNIPNI 253
LLF R E A +MKNIL + E SG+AI+ +KS I+ SRN + +I I
Sbjct: 279 LLFEGAPLRSLRVEEHHAQIMKNILILYEEDSGKAISLRKS*IYCSRNVPDILKTSITYI 458
Query: 254 LQVQAVLGTGKYLGLPSMTGRSKKATFNFIKDRIWKK 290
L VQ +LGT KYLGLPSM GR + TF+ IK +W+K
Sbjct: 459 LGVQFMLGTCKYLGLPSMIGRDRTTTFSSIKGGVWQK 569
Score = 77.8 bits (190), Expect(2) = 3e-21
Identities = 40/70 (57%), Positives = 46/70 (65%)
Frame = +2
Query: 123 MCVETVDYSVIVNEEIAGPIIHGRGLRQGDPLSTYLFIICAEGLSSLIRKAEASGTIHGV 182
MCVE+ DY V+VN + PII RGL+QGD LS Y+FIIC EGLS LI A+ G HG
Sbjct: 101 MCVESNDYYVLVNNDAVDPIIPSRGLQQGDHLSPYIFIICVEGLSFLIPHAKERGDTHGT 280
Query: 183 KICNNAPIVS 192
I AP VS
Sbjct: 281 SI*RGAPPVS 310
Score = 43.1 bits (100), Expect(2) = 3e-21
Identities = 17/32 (53%), Positives = 26/32 (81%)
Frame = +1
Query: 90 LDISKAYDRIDWNYLKCVMLKMGFSIQWVKWI 121
L ISK Y+R+D +YLK +M+KMGF+ +W+ W+
Sbjct: 1 LYISKVYNRVD*DYLKEIMIKMGFNNRWIYWM 96
Score = 34.3 bits (77), Expect = 0.21
Identities = 16/29 (55%), Positives = 20/29 (68%)
Frame = +1
Query: 280 FNFIKDRIWKKINSWSSKCLSKAGREVLM 308
F ++ KKINSWS CLSKA REV++
Sbjct: 538 FLLLRVECGKKINSWSDICLSKACREVMI 624
>TC79628 similar to GP|17104799|gb|AAL34288.1 putative 2-nitropropane
dioxygenase {Arabidopsis thaliana}, partial (97%)
Length = 1560
Score = 64.7 bits (156), Expect(2) = 1e-18
Identities = 33/70 (47%), Positives = 42/70 (59%)
Frame = +1
Query: 749 ICIRDDEGAFVIARTKCLTPKCDVHIGEALGLLTALQWVHELQLGPIEFELDSKKVVDSF 808
+CI D G +V+ RT P C V I +ALGL ALQWV +L + I+F LDSK VVD
Sbjct: 1162 MCI*DATGGYVLGRTTWFAPLCSVDIDKALGLRDALQWVADLGMDNIDFSLDSKLVVDVV 1341
Query: 809 GANRHDSTEF 818
+N +T F
Sbjct: 1342 SSNNSSNTGF 1371
Score = 47.4 bits (111), Expect(2) = 1e-18
Identities = 25/62 (40%), Positives = 35/62 (56%)
Frame = +3
Query: 819 GAIINDCKTLFNHFYENSSVEFVRRQANEAAHELAKAATLSASFHILVTPPHCIEHILIN 878
G+IIN C + S VEF RRQAN H LA+ TL AS + + P CI +++ N
Sbjct: 1374 GSIINYCISYLVLILPISKVEFNRRQANGMTHGLAQTTTLEASSQLFIDLPPCINNLVSN 1553
Query: 879 DM 880
++
Sbjct: 1554 EI 1559
>AL382241
Length = 372
Score = 90.5 bits (223), Expect = 3e-18
Identities = 47/106 (44%), Positives = 62/106 (58%)
Frame = +1
Query: 775 GEALGLLTALQWVHELQLGPIEFELDSKKVVDSFGANRHDSTEFGAIINDCKTLFNHFYE 834
G L +L W+ + ++F LD K+VVD ++ DS+EFG II CK L +
Sbjct: 37 GRLLVYTPSLDWLSNQRFDNVDFVLDCKRVVDCINSSLDDSSEFGCIITACKQLLVDRFH 216
Query: 835 NSSVEFVRRQANEAAHELAKAATLSASFHILVTPPHCIEHILINDM 880
NS VEF RRQAN AHELA+A + S H++ P CI HIL N+M
Sbjct: 217 NSHVEFSRRQANRVAHELAQATLSNPSRHVIDDVPTCI*HILANEM 354
>AJ497561 weakly similar to GP|10140689|gb putative non-LTR retroelement
reverse transcriptase {Oryza sativa (japonica
cultivar-group)}, partial (2%)
Length = 621
Score = 74.3 bits (181), Expect(2) = 6e-18
Identities = 32/70 (45%), Positives = 48/70 (67%)
Frame = +2
Query: 395 TLIARIYKARYFPRCDFLQSSLGHNPSYVWRSLCNSKFILKAGSRWRIGDGDTISIWNNK 454
+L+++I K +YFP+ DF ++LGHNPS+ WRSL +++ +L G RW IGDG I++ +
Sbjct: 410 SLLSKILKFKYFPQWDFSYANLGHNPSFTWRSLLSTQSLLTLGHRWMIGDGSQINVSSMS 589
Query: 455 WIAGDITLIP 464
WI TL P
Sbjct: 590 WIRNRPTLRP 619
Score = 35.4 bits (80), Expect(2) = 6e-18
Identities = 15/37 (40%), Positives = 26/37 (69%)
Frame = +3
Query: 357 WDRLAAHKNDGGMSFKSLPVFNLAMLGKQGWRSMTNP 393
++ LA HK+ GG+ ++ FN++MLGKQ W+ ++ P
Sbjct: 303 FNNLARHKSVGGLGLQNQE-FNISMLGKQRWKLLSQP 410
>AW980456
Length = 779
Score = 88.6 bits (218), Expect = 1e-17
Identities = 41/80 (51%), Positives = 53/80 (66%)
Frame = -2
Query: 505 IIKTSLYTSIKEDRFVWNKENNGEYSVRSAYRLCMQKLLDISEFKVLGAWDYIWNLKVPP 564
I+ T L + DR +W E +G+Y V+SAYR C+++L D S G W IW LKVPP
Sbjct: 244 IMSTPLISHA*LDRLIWKDEKHGKYYVKSAYRFCVEELFDSSYLHRPGNWSGIWKLKVPP 65
Query: 565 KVKNLVWRVARNILPTRMRL 584
KV+NLVWR+ R LPTR+RL
Sbjct: 64 KVQNLVWRMCRGCLPTRIRL 5
>BG587113 weakly similar to PIR|A84888|A8 hypothetical protein At2g45230
[imported] - Arabidopsis thaliana, partial (10%)
Length = 767
Score = 83.6 bits (205), Expect = 3e-16
Identities = 57/229 (24%), Positives = 98/229 (41%), Gaps = 18/229 (7%)
Frame = -3
Query: 412 LQSSLGHNPSYVWRSLCNSKFILKAGSRWRIGDGDTISIWNNK--WIAGDITLIP----- 464
L + LG SY WRS+ +++ ++K G++ IG+G+ +IW + W +T P
Sbjct: 762 LNAPLGSWASYAWRSIHSAQHLIKQGAKVIIGNGENTNIWEREMAWKLTCVTNHPNKHSS 583
Query: 465 -----PTVGDFHLSHLRVSHCMLQHHKMWDVPFLETIFDQQTVAAIIKTSLYTSIKEDRF 519
PT+ + R M + + + +IF + T I+ I ED +
Sbjct: 582 RAY*APTL--YGYEGCRSDDPMRRER---NANLINSIFPEGTRRKILSIHPQGPIGEDSY 418
Query: 520 VWNKENNGEYSVRSAYRLCMQKLL------DISEFKVLGAWDYIWNLKVPPKVKNLVWRV 573
W +G YSV+S Y + + + + + + +W PKV++ +WR
Sbjct: 417 SWEYSKSGHYSVKSGYYVQTNIIAAANQRGTVDQPSLDDLYQRVWKYNTSPKVRHFLWRC 238
Query: 574 ARNILPTRMRLREKIVNCPPHCVLCNSADEDSLHLFFNCPSSSNIWNLS 622
N LPT +R + ++ C C E H+ F CP + IW S
Sbjct: 237 ISNSLPTAANMRSRHISKDGSCSRCGMESETVNHILFQCPYARLIWATS 91
>AL382240
Length = 320
Score = 77.4 bits (189), Expect = 2e-14
Identities = 38/103 (36%), Positives = 59/103 (56%), Gaps = 1/103 (0%)
Frame = +3
Query: 683 VLERASSLLYDWNAARNARLNNNVVNGDGEHSQST-RVIKWSKPNRGRMKCNIDASFPNN 741
V +RA+ + +W + R ++N + + S W KP G KCNIDASF +
Sbjct: 9 VFQRATHVFEEWRTTQRIRSSSNTQSTTPQVQPSRPEEDVWKKPAPGXYKCNIDASFSTS 188
Query: 742 ENRIGIGICIRDDEGAFVIARTKCLTPKCDVHIGEALGLLTAL 784
N +G+G+C+RD++ AFV+ART+ P CD+ +G L + T L
Sbjct: 189 FNIVGLGMCLRDEDDAFVLARTEWFAPLCDIDVGGGLLVYTHL 317
>BG585866
Length = 828
Score = 77.4 bits (189), Expect = 2e-14
Identities = 53/204 (25%), Positives = 83/204 (39%), Gaps = 4/204 (1%)
Frame = +3
Query: 427 LCNSKFILKAGSRWRIGDGDTISIWNNKWIAGDITLIPPTVGDFHLSHLRVSHCML---Q 483
+ K +LK+G WR G G++ S W W + + D H HL V Q
Sbjct: 84 IIRDKNVLKSGYTWRAGSGNS-SFWYTNWSSLGLLGTQAPFVDIHDLHLTVKDVFTTGGQ 260
Query: 484 HHKMWDVPFLETIFDQQTVAAIIKTSL-YTSIKEDRFVWNKENNGEYSVRSAYRLCMQKL 542
H + L TI I T L + + D ++W +NG Y+ +S Y + +
Sbjct: 261 HTQS-----LYTILPTDIAEVINNTHLNFNASIGDAYIWPHNSNGVYTAKSGYSWILSQT 425
Query: 543 LDISEFKVLGAWDYIWNLKVPPKVKNLVWRVARNILPTRMRLREKIVNCPPHCVLCNSAD 602
++ +W +IW LK+P K K +W N +PT L + + C C +
Sbjct: 426 ETVNYNN--SSWSWIWRLKIPEKYKFFLWLACHNAVPTLSLLNHRNMVNSAICSRCGEHE 599
Query: 603 EDSLHLFFNCPSSSNIWNLSNLSN 626
E H +C S IW+ S+
Sbjct: 600 ESFFHCVRDCRFSKIIWHKIGFSS 671
>TC93101
Length = 675
Score = 75.5 bits (184), Expect = 8e-14
Identities = 54/215 (25%), Positives = 95/215 (44%), Gaps = 8/215 (3%)
Frame = -1
Query: 578 LPTRMRLREKIVNCPPHCVLCNSADEDSLHLFFNCPSSSNIWNLSNLSNDVKIVVSQGLN 637
LP R L ++ VNCPP C C E + H+F +C + +W S LS ++ + +N
Sbjct: 663 LPVRXELNKRGVNCPPLCPRCYFNLETTNHIFMSCERTQRVWFGSQLS--IRFPDNSTIN 490
Query: 638 SSAVIFHLLQVLSAGESALFATILWSIWKQRNNKIWNDVTDAQNFVLERASSLLYDWNAA 697
S +F + + + I +SIW RN I+ + +++ +++ A + + + A
Sbjct: 489 FSDWLFDAISNQTEEIIIKISAITYSIWHARNKAIFENQFVSEDTIIQ*AQNSILAYEQA 310
Query: 698 RNARLNNNVV--------NGDGEHSQSTRVIKWSKPNRGRMKCNIDASFPNNENRIGIGI 749
N N+V N + +S +W KP +K N DA+ + R G+G
Sbjct: 309 TIKPQNPNIVLSSLSATSNTNTTRRRSNVRSRWQKPLNNILKANCDANL-QVQGRWGLGC 133
Query: 750 CIRDDEGAFVIARTKCLTPKCDVHIGEALGLLTAL 784
IR+ +G + T C+ E +L A+
Sbjct: 132 IIRNADGEAKVTATWCINGFDCAATAETYAILAAM 28
>BG586862
Length = 804
Score = 73.9 bits (180), Expect = 2e-13
Identities = 64/251 (25%), Positives = 108/251 (42%), Gaps = 6/251 (2%)
Frame = -1
Query: 557 IWNLKVPPKVKNLVWRVARNILPTRMRLREKIVNCPPHCVLCNSADEDSLHLFFNCPSSS 616
+W +K P+ K+ +WR+ N LP + L ++ + C C C S E HLF NC +
Sbjct: 654 VWGIKTIPRHKSFLWRLLHNALPVKDELHKRGIRCSLLCPRCESKIETVQHLFLNCEVTQ 475
Query: 617 NIWNLSNLSNDVKIVVSQGLNSSAVI-FH---LLQVLSAGESALFA--TILWSIWKQRNN 670
W S L + +SS V+ FH +L E + A +L+SIW RN
Sbjct: 474 KEWFGSQLGIN--------FHSSGVLHFHDWITNFILKNDEETIIALTALLYSIWHARNQ 319
Query: 671 KIWNDVTDAQNFVLERASSLLYDWNAARNARLNNNVVNGDGEHSQSTRVIKWSKPNRGRM 730
K++ ++ + V++RASS L+ + + A+++++V+ + S S W
Sbjct: 318 KVFENIDVPGDVVIQRASSSLHSF---KMAQVSDSVLPSNAIPSYSL----W-------- 184
Query: 731 KCNIDASFPNNENRIGIGICIRDDEGAFVIARTKCLTPKCDVHIGEALGLLTALQWVHEL 790
GIG+ R+ EG + + T EA G+ A+ + +
Sbjct: 183 ---------------GIGVVARNCEGLAMASGTWLRHGIPCATTAEAWGIYQAMVFAGDC 49
Query: 791 QLGPIEFELDS 801
EFE D+
Sbjct: 48 GFSKFEFESDN 16
>BQ149324
Length = 374
Score = 72.0 bits (175), Expect = 9e-13
Identities = 37/91 (40%), Positives = 54/91 (58%)
Frame = +1
Query: 583 RLREKIVNCPPHCVLCNSADEDSLHLFFNCPSSSNIWNLSNLSNDVKIVVSQGLNSSAVI 642
+++E V HCV A LHL F C SS N+W++ + + I++ Q ++S +I
Sbjct: 25 KIKESHVLWIVHCV--RWAVRIHLHLIFQCSSSLNVWSMLPFLSTISILLQQDMDSKNII 198
Query: 643 FHLLQVLSAGESALFATILWSIWKQRNNKIW 673
F L LS ++ALF +LWSI KQRNNK+W
Sbjct: 199 FKALHDLSNEDAALFCCVLWSI*KQRNNKVW 291
Score = 37.7 bits (86), Expect = 0.019
Identities = 15/28 (53%), Positives = 19/28 (67%)
Frame = +3
Query: 578 LPTRMRLREKIVNCPPHCVLCNSADEDS 605
LPTR+RL++K V CP C LC ED+
Sbjct: 3 LPTRVRLKDKRVTCPMDCTLCTVGSEDT 86
>BG447966 weakly similar to PIR|G96509|G96 protein F27F5.21 [imported] -
Arabidopsis thaliana, partial (7%)
Length = 687
Score = 71.2 bits (173), Expect = 2e-12
Identities = 38/67 (56%), Positives = 45/67 (66%)
Frame = +2
Query: 2 TNIALIPKGDMQVSMKDWRPIALCNVVYKIAAKVLANRLKRVLDKCISDNQSPFVPGRSI 61
T I LIPKG + KD+RPI+LCNVV KI KV+ANR+K+ L I QS FV GR I
Sbjct: 434 TFIVLIPKGKNPNTPKDYRPISLCNVVMKIITKVIANRVKQTLPDVIDVEQSAFVQGRLI 613
Query: 62 LDNAMAA 68
DNA+ A
Sbjct: 614 TDNALIA 634
>BQ148771
Length = 680
Score = 68.6 bits (166), Expect = 1e-11
Identities = 41/155 (26%), Positives = 79/155 (50%), Gaps = 2/155 (1%)
Frame = -3
Query: 275 SKKATFNFIKDRIWKKINSWSSKCLSKAGREVLMKSMLQSIPTYFMSIFTLPTSLCDEIE 334
+KK+ F+ + ++ + +W + LS A R L KS+++++P Y M +P + +EI+
Sbjct: 618 TKKSRFS-VYYQVHVMLANWKANHLSLARRVTLAKSVIEAVPLYPMMTTIIPKACIEEIQ 442
Query: 335 KMLNSFWWGHSGTQGRGLHWLSWDRLAAHKNDGGMSFKSLPVFNLAMLGKQGWRSMTNPD 394
K+ F WG + R H + W+ ++ K G+ + L V N A + K GW + +
Sbjct: 441 KLQRKFVWGDTEV-SRRYHAVGWETMSKPKTIYGLGLRRLDVMNKACIMKLGWSIYSGSN 265
Query: 395 TLIARIYKARYFPRCDFLQSSLGHNP--SYVWRSL 427
+L + + +Y R + L+ P S +W++L
Sbjct: 264 SLCTEVMRGKY-QRSESLEEIFLEKPTDSSLWKAL 163
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.322 0.136 0.426
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 33,191,420
Number of Sequences: 36976
Number of extensions: 529558
Number of successful extensions: 2643
Number of sequences better than 10.0: 94
Number of HSP's better than 10.0 without gapping: 2585
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2619
length of query: 881
length of database: 9,014,727
effective HSP length: 105
effective length of query: 776
effective length of database: 5,132,247
effective search space: 3982623672
effective search space used: 3982623672
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 63 (28.9 bits)
Medicago: description of AC138579.1


