

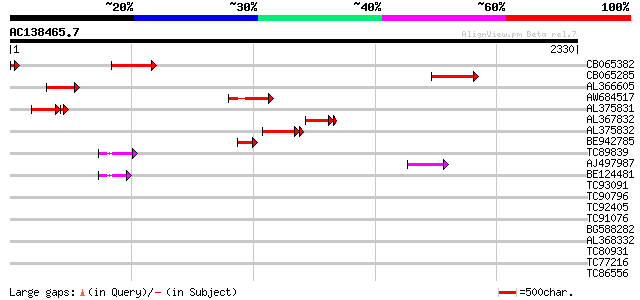
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC138465.7 + phase: 0 /pseudo
(2330 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
CB065382 similar to PIR|T09671|T096 RPE15 protein - alfalfa (fra... 308 2e-83
CB065285 weakly similar to PIR|A84500|A84 probable retroelement ... 301 1e-81
AL366605 270 6e-72
AW684517 similar to GP|9927273|dbj Similar to Arabidopsis thalia... 265 2e-70
AL375831 200 1e-66
AL367832 weakly similar to GP|14091845|gb Putative retroelement ... 208 1e-53
AL375832 200 1e-50
BE942785 145 1e-34
TC89839 homologue to SP|P23466|CYAA_SACKL Adenylate cyclase (EC ... 76 2e-13
AJ497987 weakly similar to GP|9927273|dbj Similar to Arabidopsis... 74 5e-13
BE124481 57 6e-08
TC93091 similar to GP|4115538|dbj|BAA36412.1 UDP-glycose:flavono... 37 0.069
TC90796 similar to GP|3395431|gb|AAC28763.1| unknown protein {Ar... 37 0.12
TC92405 similar to GP|11994325|dbj|BAB02284. gene_id:MLM24.13~un... 37 0.12
TC91076 similar to PIR|H96747|H96747 unknown protein T10D10.14 [... 35 0.44
BG588282 similar to PIR|D84731|D847 probable uclacyanin I [impor... 35 0.44
AL368332 weakly similar to GP|21322711|e pherophorin-dz1 protein... 34 0.58
TC80931 similar to GP|14190783|gb|AAF65166.2 putative phloem tra... 34 0.76
TC77216 similar to PIR|G85040|G85040 probable xyloglucan endotra... 33 0.99
TC86556 similar to PIR|T07598|T07598 proline-rich protein GPP1 -... 33 1.3
>CB065382 similar to PIR|T09671|T096 RPE15 protein - alfalfa (fragment),
partial (9%)
Length = 624
Score = 308 bits (788), Expect = 2e-83
Identities = 151/189 (79%), Positives = 162/189 (84%), Gaps = 1/189 (0%)
Frame = -2
Query: 417 QPQVPVNAVVQQMQQQPPV-QQQQHQQARPTFPPIPMLYAELLPTLLLRGHCTTRHGKPP 475
+P +P PPV QQ+Q QQARPTFP IPMLYAE LPTLLLRGHCT R GKPP
Sbjct: 569 RPMIP*LLC*PPRNNHPPVHQQRQ*QQARPTFPLIPMLYAE*LPTLLLRGHCTIRQGKPP 390
Query: 476 PDPLLLRFRSDLKCDFHQGALGHDVEGCYTLKYIVKKLIDQGKLTFENNVPYVLDNPLPN 535
PDPL RFRSDLKCDFHQGALGHDVEGCY LK+IVKKLI+QGKLTFENNVP+VLDNPLPN
Sbjct: 389 PDPLPPRFRSDLKCDFHQGALGHDVEGCYALKHIVKKLINQGKLTFENNVPHVLDNPLPN 210
Query: 536 HAAVNMIEVYEEAPRLDVHNIATPLVPLHIKLCQASLFDHDHASCPE*FHNTLGCYVVQN 595
HAAVNMIEVYEEAP LDV N+ TPLVPLHIKLCQASLFDHDHA+C E*F+N LGC VVQN
Sbjct: 209 HAAVNMIEVYEEAPGLDVRNVTTPLVPLHIKLCQASLFDHDHANCQE*FYNPLGCCVVQN 30
Query: 596 EIQSLMNDN 604
+IQSLMN+N
Sbjct: 29 DIQSLMNNN 3
Score = 67.0 bits (162), Expect = 8e-11
Identities = 34/40 (85%), Positives = 36/40 (90%)
Frame = -1
Query: 1 NAQLRTELASLREELAKANDTMTALLAAQEQSATAIPVAT 40
NAQLRTELAS+REELAKANDTMTALLAAQEQS+T P T
Sbjct: 618 NAQLRTELASMREELAKANDTMTALLAAQEQSSTGSPTTT 499
>CB065285 weakly similar to PIR|A84500|A84 probable retroelement gag/pol
polyprotein [imported] - Arabidopsis thaliana, partial
(2%)
Length = 592
Score = 301 bits (772), Expect = 1e-81
Identities = 164/195 (84%), Positives = 170/195 (87%)
Frame = -3
Query: 1732 NQKIAKNR*LVKVQIPIASGG*SLMVLSMLMAKELGQSLYPRRGITFLLLPGFCSNVQTI 1791
NQK AKNR*LVKVQIPIA+G *SLMVLSMLM KEL QSLYPRRGIT LL PGFCSNVQTI
Sbjct: 590 NQKTAKNR*LVKVQIPIANGV*SLMVLSMLMVKELEQSLYPRRGITSLLPPGFCSNVQTI 411
Query: 1792 WLSMKHVSLGSRKQLI*ESNTSTSMEILHLSSTR*RVNGRPTMPS*FLIVITRDVC*HIL 1851
WLSMKHVSLGSRKQL *ESNTSTSMEILHLSSTR RVNGRPTMP *FL R VC* IL
Sbjct: 410 WLSMKHVSLGSRKQLT*ESNTSTSMEILHLSSTRSRVNGRPTMPI*FLTATMRGVC*PIL 231
Query: 1852 QRLSYTIFLVMRTKWLMLLLLYPPCFE*IIGMMCQ*SKYNALKDLLMCLLLKI*SVGLMK 1911
Q+LS TIFLV RTKWLML LLYPPCFE* IGMMCQ*SK NALKDL MCLLL +*S+ +K
Sbjct: 230 QKLSCTIFLVTRTKWLMLWLLYPPCFE*TIGMMCQ*SKCNALKDLHMCLLLGM*SIRPVK 51
Query: 1912 MWLTINPGITTSSSS 1926
MWLTINPG TTS+SS
Sbjct: 50 MWLTINPGTTTSNSS 6
>AL366605
Length = 422
Score = 270 bits (689), Expect = 6e-72
Identities = 128/139 (92%), Positives = 132/139 (94%)
Frame = -3
Query: 149 EIKANRGNGDSVKTHDLCLVPKVDVPKKFKIPEFDRYNGLTCPQNHIIKYVRKMGNYSDN 208
EIKANRGN DS KT DLCLV KVDVPKKFKIP+FDRYNGLTCPQNHIIKYVRKMGNY DN
Sbjct: 417 EIKANRGNADSFKTQDLCLVSKVDVPKKFKIPDFDRYNGLTCPQNHIIKYVRKMGNYKDN 238
Query: 209 DSLMIHCFQDSLMEDAAEWYTSLSKDDVHTFDELAAAFKSHYGFNTRLKPNREFLISLSQ 268
DSLMIHCFQDSLMEDAAEWYTSLSK+D+HTFDELAAAFKSHYGFNTRLKPNREFL SLSQ
Sbjct: 237 DSLMIHCFQDSLMEDAAEWYTSLSKNDIHTFDELAAAFKSHYGFNTRLKPNREFLRSLSQ 58
Query: 269 KKDESFREYAQIWRGAAAR 287
KK+ESFREYAQ WRGAAAR
Sbjct: 57 KKEESFREYAQRWRGAAAR 1
>AW684517 similar to GP|9927273|dbj Similar to Arabidopsis thaliana chromosome
II BAC F26H6; putative retroelement pol polyprotein,
partial (1%)
Length = 488
Score = 265 bits (677), Expect = 2e-70
Identities = 138/187 (73%), Positives = 147/187 (77%)
Frame = +1
Query: 898 ITFQVMDINASYSCLLGRPWIHDAGAVTSTLHKKLKFVKNGKLVTVHGEEAYLVSQLSSF 957
ITFQVMDINASYSCLLGRPWIHDAGAVTSTLH+KLKFVKNG
Sbjct: 1 ITFQVMDINASYSCLLGRPWIHDAGAVTSTLHQKLKFVKNG------------------- 123
Query: 958 SCIEAGSAEGTAFQGLTIEGTELKKTGTAMASLKDAQKAVQEGQAAGWGKLIQLRENKHK 1017
SAEGTAFQGL++EG E KK G AMASLKDAQKAVQEGQAA WGKLIQL ENK K
Sbjct: 124 ------SAEGTAFQGLSMEGAEPKKVGAAMASLKDAQKAVQEGQAADWGKLIQLCENKRK 285
Query: 1018 EGLGFSPTSGVST*TFHSAGFVNAITEEAAGFGPRPVFVIPGGIARDWDAIDIPTIMHVS 1077
EGL FSPTSGVST TFHSAGFVN + EE A F PRP+FVIPGGIA+DWDA+D+P+IMHVS
Sbjct: 286 EGLRFSPTSGVSTGTFHSAGFVNTLAEEVARFVPRPLFVIPGGIAKDWDAVDVPSIMHVS 465
Query: 1078 E*CVLLL 1084
*CVLLL
Sbjct: 466 X*CVLLL 486
>AL375831
Length = 467
Score = 200 bits (509), Expect(2) = 1e-66
Identities = 96/119 (80%), Positives = 101/119 (84%)
Frame = +2
Query: 89 VPGTNTISINTTLPQTTATVTEPLVHAISQGVNINAHHGSIPVIKTMEERMEELAKELRR 148
+P TN SI TLPQTTA VTEPLVH + QG+NIN H SIPV KTMEE MEELAKELR
Sbjct: 8 IPATNAASIIATLPQTTAAVTEPLVHTLPQGININTQHRSIPVTKTMEEMMEELAKELRH 187
Query: 149 EIKANRGNGDSVKTHDLCLVPKVDVPKKFKIPEFDRYNGLTCPQNHIIKYVRKMGNYSD 207
EI+ANRGN DSVKT DLCLV KVDVPKKFKIP+FDRYNGLTCPQNHIIKYVRKMGNY D
Sbjct: 188 EIQANRGNADSVKTQDLCLVSKVDVPKKFKIPDFDRYNGLTCPQNHIIKYVRKMGNYKD 364
Score = 73.9 bits (180), Expect(2) = 1e-66
Identities = 32/34 (94%), Positives = 34/34 (99%)
Frame = +3
Query: 208 NDSLMIHCFQDSLMEDAAEWYTSLSKDDVHTFDE 241
NDSLMIHCFQDSLMEDAAEWYTSLSK+D+HTFDE
Sbjct: 366 NDSLMIHCFQDSLMEDAAEWYTSLSKNDIHTFDE 467
>AL367832 weakly similar to GP|14091845|gb Putative retroelement {Oryza
sativa}, partial (2%)
Length = 384
Score = 208 bits (530), Expect(2) = 1e-53
Identities = 106/118 (89%), Positives = 111/118 (93%)
Frame = -3
Query: 1214 RKESHSASSGRD*AYQHRYRREQARNQGRCCSKGRG*KEDHPAPPRIPGYFCMVVRRYAR 1273
RKESHSAS GRD*AYQ RYR EQAR+QGRCCS+GRG*KED PAPPRIPGYFCM+V RYAR
Sbjct: 382 RKESHSASPGRD*AYQPRYRGEQARDQGRCCSRGRG*KEDLPAPPRIPGYFCMLV*RYAR 203
Query: 1274 SRS*DCGTSDPHKA*MSSRQAEIEKDSSRYGSQD*E*GSKAD*CGFSHDSRVS*MGCQ 1331
SRS*+CGTSDP+KA*MSSRQ EIEKDSSRYGSQD*E*GSKAD*CGFSHDS VS*MGCQ
Sbjct: 202 SRS*NCGTSDPNKA*MSSRQVEIEKDSSRYGSQD*E*GSKAD*CGFSHDS*VS*MGCQ 29
Score = 21.9 bits (45), Expect(2) = 1e-53
Identities = 9/12 (75%), Positives = 9/12 (75%)
Frame = -2
Query: 1329 GCQPLYLCQRRM 1340
G LYLCQRRM
Sbjct: 38 GLPILYLCQRRM 3
>AL375832
Length = 535
Score = 200 bits (509), Expect(2) = 1e-50
Identities = 113/163 (69%), Positives = 122/163 (74%), Gaps = 9/163 (5%)
Frame = +3
Query: 1038 FVNAITEEAAGFGPRPVFVIPGGIARDWDAIDIPTIMHVSE*CVLLLKCFVFPNNNPLAL 1097
FVNAI+EEA G G RP FV PGGIA DWDAIDIP+IMHVSE*CVLLL FVF NNNPLAL
Sbjct: 3 FVNAISEEATGSGLRPAFVTPGGIASDWDAIDIPSIMHVSE*CVLLL*RFVFKNNNPLAL 182
Query: 1098 PEAKVIFSVRACLFRHRRY**KS-SFLSRLLFFSTFFPWK*W*CQKKPKHLYFLINFKNL 1156
P+A+VIFSVRACLFRH R K+ F FFS FF K W*CQKKPK LYFLINFKNL
Sbjct: 183 PKARVIFSVRACLFRHCRLMNKNCRFFHDCFFFSAFFLEK-W*CQKKPKRLYFLINFKNL 359
Query: 1157 H--------KNLFLKSSNHFTQIEP**TRWAR*PYLSSEF*IP 1191
H K LFLKSSNH+ Q+EP**TR +* Y SS+ * P
Sbjct: 360 HEKKKIQKTKKLFLKSSNHYMQVEP**TR*TQ*FYASSKL*FP 488
Score = 20.0 bits (40), Expect(2) = 1e-50
Identities = 11/18 (61%), Positives = 14/18 (77%)
Frame = +2
Query: 1190 IPGV*SRR*GR**HTV*D 1207
IPG+*+ G **+TV*D
Sbjct: 482 IPGI*TEDEGG**YTV*D 535
>BE942785
Length = 460
Score = 145 bits (367), Expect = 1e-34
Identities = 70/82 (85%), Positives = 78/82 (94%)
Frame = -2
Query: 937 NGKLVTVHGEEAYLVSQLSSFSCIEAGSAEGTAFQGLTIEGTELKKTGTAMASLKDAQKA 996
+G LVT+HGEEAYL+SQLSSFSCIEAGSAEGTAFQGLT+EGTE K+ GTAMASLKDAQ+A
Sbjct: 378 SGXLVTIHGEEAYLISQLSSFSCIEAGSAEGTAFQGLTVEGTEPKRDGTAMASLKDAQRA 199
Query: 997 VQEGQAAGWGKLIQLRENKHKE 1018
VQE QAAGWG+LIQLRENKHK+
Sbjct: 198 VQESQAAGWGRLIQLRENKHKD 133
>TC89839 homologue to SP|P23466|CYAA_SACKL Adenylate cyclase (EC 4.6.1.1)
(ATP pyrophosphate-lyase) (Adenylyl cyclase). [Yeast],
partial (0%)
Length = 665
Score = 75.9 bits (185), Expect = 2e-13
Identities = 55/158 (34%), Positives = 76/158 (47%)
Frame = +3
Query: 366 EVGMVSAGAGQSMATVAPINAAQLPPSYPYAPYSQHPFFPPFYHQYPLPSGQPQVPVNAV 425
EV + A + + V+ + A PP P S + P Q P + Q P ++
Sbjct: 156 EVTTLRAEVEKLTSLVSSLMATNDPPLVQQRPQSP---YQPMCPQKP----RQQAPRQSI 314
Query: 426 VQQMQQQPPVQQQQHQQARPTFPPIPMLYAELLPTLLLRGHCTTRHGKPPPDPLLLRFRS 485
Q Q + Q Q Q+A PIP+ YA+LLP LL + T P+ L +R
Sbjct: 315 PQNQVPQKFIPQNQVQKASQC-DPIPVKYADLLPILLKKNLIQTLPLPRVPNSLPPWYRP 491
Query: 486 DLKCDFHQGALGHDVEGCYTLKYIVKKLIDQGKLTFEN 523
DL C FHQGA GHD E CY LK V+KLI+ +F++
Sbjct: 492 DLNCVFHQGAPGHDTEQCYPLKEEVQKLIENNVWSFDD 605
>AJ497987 weakly similar to GP|9927273|dbj Similar to Arabidopsis thaliana
chromosome II BAC F26H6; putative retroelement pol
polyprotein, partial (1%)
Length = 636
Score = 74.3 bits (181), Expect = 5e-13
Identities = 68/169 (40%), Positives = 80/169 (47%)
Frame = -3
Query: 1636 RLVVLWPGPPNACVIIW*IIQLG*YPEWIR*SISLRKLLLLGRLHAGRCSCLNMILCSKL 1695
R VVL PG P+A VII I LG Y +WI+ S LR L LH GRC NMI +
Sbjct: 634 RHVVL*PGLPSASVII*LITLLGWYLKWIQSSTYLRSPL*QEGLHGGRCYYRNMISSTVP 455
Query: 1696 KRQSKVAFLPIILPTNLLMITNQLSSISPMKRSCI*NQKIAKNR*LVKVQIPIASGG*SL 1755
+RQ K FL I TN L + S MKR CI* KI + KV I I G * L
Sbjct: 454 RRQLKAVFLLITWLTNHLKTIDLSSLTFLMKRLCI*R*KIVTSHYSEKVLIQIQCGV*YL 275
Query: 1756 MVLSMLMAKELGQSLYPRRGITFLLLPGFCSNVQTIWLSMKHVSLGSRK 1804
M LSM A GQ + +T L +T L+ K S S +
Sbjct: 274 MGLSMYTATA*GQYSLLLKVLTSLSQQDSGLTARTTSLNTKPASWASTR 128
>BE124481
Length = 538
Score = 57.4 bits (137), Expect = 6e-08
Identities = 45/134 (33%), Positives = 61/134 (44%)
Frame = +3
Query: 366 EVGMVSAGAGQSMATVAPINAAQLPPSYPYAPYSQHPFFPPFYHQYPLPSGQPQVPVNAV 425
EV + A + + V+ + A PP P S + P Q P + Q P ++
Sbjct: 159 EVTTLRAEVEKLTSLVSSLMATNDPPLVQQRPQSP---YQPMCPQKP----RQQAPRQSI 317
Query: 426 VQQMQQQPPVQQQQHQQARPTFPPIPMLYAELLPTLLLRGHCTTRHGKPPPDPLLLRFRS 485
Q Q + Q Q Q+A PIP+ YA+LLP LL + T P+ L +R
Sbjct: 318 PQNQVPQKFIPQNQVQKASQC-DPIPVKYADLLPILLKKNLIQTLPLPRVPNSLPPWYRP 494
Query: 486 DLKCDFHQGALGHD 499
DL C FHQGA GHD
Sbjct: 495 DLNCVFHQGAPGHD 536
>TC93091 similar to GP|4115538|dbj|BAA36412.1 UDP-glycose:flavonoid
glycosyltransferase {Vigna mungo}, partial (37%)
Length = 845
Score = 37.4 bits (85), Expect = 0.069
Identities = 29/105 (27%), Positives = 45/105 (42%), Gaps = 16/105 (15%)
Frame = -3
Query: 369 MVSAGAGQSMATVAPINAAQL---PPSYPYAPYSQHPFFPPFYHQY-----------PLP 414
++S Q +T++P + L P P+AP Q P +PP H Y L
Sbjct: 519 LLSLSLSQIPSTLSPSSPLPLLSQTPHLPFAPPQQPPSYPPPSHSY*HAALHAMAMQSLA 340
Query: 415 SGQPQVPV-NAVVQQMQQQPPVQQQQHQQARPT-FPPIPMLYAEL 457
QP +P ++ Q P++ +H+ PT PP P L+ L
Sbjct: 339 LQQPPLPTPSSSHSDSQTHQPIRDSEHRLVGPTSSPPGPSLWFSL 205
>TC90796 similar to GP|3395431|gb|AAC28763.1| unknown protein {Arabidopsis
thaliana}, partial (20%)
Length = 965
Score = 36.6 bits (83), Expect = 0.12
Identities = 26/66 (39%), Positives = 30/66 (45%), Gaps = 8/66 (12%)
Frame = +2
Query: 392 SYPYAPYSQHPFFPPFYHQYPLPSGQPQVPVNAVVQQMQQQ--------PPVQQQQHQQA 443
SY YAP + +P P Y+ Y P QPQ QQ QQ PP QQ Q
Sbjct: 773 SYSYAPQA-YPGNSP-YNSYVAPWAQPQSKSEFQTQQPQQHMYQPQPTTPPSPQQHMYQP 946
Query: 444 RPTFPP 449
+PT PP
Sbjct: 947 QPTTPP 964
>TC92405 similar to GP|11994325|dbj|BAB02284. gene_id:MLM24.13~unknown
protein {Arabidopsis thaliana}, partial (69%)
Length = 864
Score = 36.6 bits (83), Expect = 0.12
Identities = 32/111 (28%), Positives = 49/111 (43%), Gaps = 1/111 (0%)
Frame = +2
Query: 831 KSDMLSNVLVDTGSSLNVMPKSTLDQLSYRETPLRRSTFLVKAFDGSRKNV-LGEIDLPM 889
K+D + L D G +++ LD+L R + S F + GSR +G + LP+
Sbjct: 278 KADAAAKELEDAGGLVDLT--DNLDRLQGRWKLIYSSAFSSRTLGGSRPGPPIGRL-LPI 448
Query: 890 TIGHETFLITFQVMDINASYSCLLGRPWIHDAGAVTSTLHKKLKFVKNGKL 940
T+G I D + LG PW VT+TL K + V + K+
Sbjct: 449 TLGQVFQRIDILSKDFDNIVDLQLGAPWPLPPLEVTATLAHKFELVGSSKI 601
>TC91076 similar to PIR|H96747|H96747 unknown protein T10D10.14 [imported] -
Arabidopsis thaliana, partial (9%)
Length = 834
Score = 34.7 bits (78), Expect = 0.44
Identities = 32/104 (30%), Positives = 42/104 (39%), Gaps = 6/104 (5%)
Frame = +3
Query: 350 NASKRYGNDHHKKKETEVGMVSAGAGQSMATVAPI--NAAQLPPSYPYA-PYSQHPFFPP 406
N ++YG + E ++ G+ S+ + NA LPP P SQ
Sbjct: 270 NEMQQYGEPIPGQSNNEAAKLAGGSNASLNLPQSLVANARMLPPGNPQGLQMSQALLSGV 449
Query: 407 FYHQYPLPSGQPQVPVNAVVQQMQQQPPVQQQQH---QQARPTF 447
Q P Q AV+QQ QQQ +QQ QH QQ P F
Sbjct: 450 SMAQRPQQLDSQQ----AVLQQQQQQQQLQQNQHSLLQQQNPQF 569
>BG588282 similar to PIR|D84731|D847 probable uclacyanin I [imported] -
Arabidopsis thaliana, partial (17%)
Length = 677
Score = 34.7 bits (78), Expect = 0.44
Identities = 24/80 (30%), Positives = 41/80 (51%), Gaps = 5/80 (6%)
Frame = +3
Query: 667 PLASLAMVSNIAEGTTAALRSGRVRPPLFQKKADTPTTPPIDKAT--LTDVSPV---TKD 721
P+ S+ + AE T ++ S PLF+ + ++PT P+ +T L SP+ ++D
Sbjct: 243 PMISIIPSAAPAESTVSSAESPEASSPLFEAQVESPTLSPMIPSTEFLAPSSPIAQHSQD 422
Query: 722 VSRPSQSIEDSNLDEILRII 741
VS + S E NL + I+
Sbjct: 423 VS--ASSTEKGNLQAFISIV 476
>AL368332 weakly similar to GP|21322711|e pherophorin-dz1 protein {Volvox
carteri f. nagariensis}, partial (10%)
Length = 478
Score = 34.3 bits (77), Expect = 0.58
Identities = 24/76 (31%), Positives = 31/76 (40%), Gaps = 3/76 (3%)
Frame = +1
Query: 382 APINAAQLPPSYPYA---PYSQHPFFPPFYHQYPLPSGQPQVPVNAVVQQMQQQPPVQQQ 438
AP +++ PP P P HPF PP H +P PS P P+ ++ PP
Sbjct: 190 APSHSSPPPPPPPRKXPPPPPSHPFSPPPPHNHPPPS--PHHPIAPSPPHVRPSPP---- 351
Query: 439 QHQQARPTFPPIPMLY 454
P PP P Y
Sbjct: 352 ------PPLPPSPSPY 381
>TC80931 similar to GP|14190783|gb|AAF65166.2 putative phloem transcription
factor M1 {Apium graveolens}, partial (67%)
Length = 1193
Score = 33.9 bits (76), Expect = 0.76
Identities = 26/94 (27%), Positives = 38/94 (39%)
Frame = +1
Query: 26 LAAQEQSATAIPVATAIPIATSVIPTASTDARFAMPTGFPYGLPPFFTPSTAAGTSGTAN 85
+ +Q+Q + I A T+ TA+T++ FP TP+TAA T T+
Sbjct: 535 MQSQQQHLSTIAATDATTTTTATASTAATES-----AAFPDSTADSTTPATAAATPSTST 699
Query: 86 NGAVPGTNTISINTTLPQTTATVTEPLVHAISQG 119
A T + T TAT T A + G
Sbjct: 700 AAAFSAAATAA---TASAVTATATSTASSAATNG 792
>TC77216 similar to PIR|G85040|G85040 probable xyloglucan
endotransglycosylase [imported] - Arabidopsis thaliana,
partial (86%)
Length = 1202
Score = 33.5 bits (75), Expect = 0.99
Identities = 16/54 (29%), Positives = 29/54 (53%)
Frame = -2
Query: 566 KLCQASLFDHDHASCPE*FHNTLGCYVVQNEIQSLMNDNYLTVSDVCVIVPVFH 619
K+ +++LF HDH P H+ C + + L+ +L V ++CV++ V H
Sbjct: 748 KVHESNLFSHDHLVLPNHLHSI*NCI---HPLLGLLGKEFLCVPNLCVLLLVSH 596
>TC86556 similar to PIR|T07598|T07598 proline-rich protein GPP1 - potato,
partial (24%)
Length = 1250
Score = 33.1 bits (74), Expect = 1.3
Identities = 30/105 (28%), Positives = 41/105 (38%), Gaps = 8/105 (7%)
Frame = +3
Query: 383 PINAAQLPPSYPYA-----PYSQHPFFP--PFYHQYPLPSGQ-PQVPVNAVVQQMQQQPP 434
P+ LPP P P +HP P P Y P+P Q P VPV ++ P
Sbjct: 165 PVYHKPLPPPVPVKKPCPPPKVEHPVLPPAPVYKPPPVPKPQPPPVPV-------KKPCP 323
Query: 435 VQQQQHQQARPTFPPIPMLYAELLPTLLLRGHCTTRHGKPPPDPL 479
+ +H P PP+P+ +P L PPP P+
Sbjct: 324 PPKVEH----PVLPPVPVYKPPPVPKAL-----------PPPVPI 413
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.338 0.146 0.475
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 75,600,509
Number of Sequences: 36976
Number of extensions: 1207992
Number of successful extensions: 9823
Number of sequences better than 10.0: 59
Number of HSP's better than 10.0 without gapping: 5350
Number of HSP's successfully gapped in prelim test: 546
Number of HSP's that attempted gapping in prelim test: 3882
Number of HSP's gapped (non-prelim): 6693
length of query: 2330
length of database: 9,014,727
effective HSP length: 112
effective length of query: 2218
effective length of database: 4,873,415
effective search space: 10809234470
effective search space used: 10809234470
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.8 bits)
S2: 66 (30.0 bits)
Medicago: description of AC138465.7


