

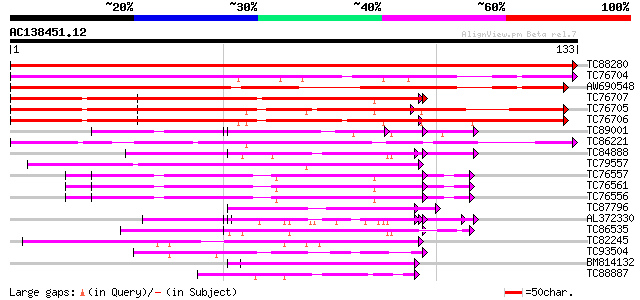
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC138451.12 - phase: 0
(133 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC88280 weakly similar to GP|7024449|dbj|BAA92155.1 glycine-rich... 288 6e-79
TC76704 homologue to SP|Q09134|GRPA_MEDFA Abscisic acid and envi... 130 1e-31
AW690548 similar to SP|Q09134|GRPA Abscisic acid and environment... 115 5e-27
TC76707 similar to SP|Q07202|CORA_MEDSA Cold and drought-regulat... 114 1e-26
TC76705 similar to SP|Q07202|CORA_MEDSA Cold and drought-regulat... 111 9e-26
TC76706 similar to SP|Q07202|CORA_MEDSA Cold and drought-regulat... 108 6e-25
TC89001 similar to GP|5726567|gb|AAD48471.1| glycine-rich RNA-bi... 57 2e-09
TC86221 similar to PIR|T09608|T09608 environmental stress-induce... 57 3e-09
TC84888 similar to GP|21322752|dbj|BAB78536. cold shock protein-... 55 1e-08
TC79557 similar to GP|12039289|gb|AAG46079.1 hypothetical protei... 54 2e-08
TC76557 similar to PIR|S71779|S71779 glycine-rich RNA-binding pr... 52 5e-08
TC76561 similar to PIR|S71779|S71779 glycine-rich RNA-binding pr... 52 5e-08
TC76556 similar to PIR|S71779|S71779 glycine-rich RNA-binding pr... 52 5e-08
TC87796 weakly similar to SP|P03211|EBN1_EBV EBNA-1 nuclear prot... 51 1e-07
AL372330 similar to GP|21322752|dbj cold shock protein-1 {Tritic... 51 2e-07
TC86535 similar to GP|9758076|dbj|BAB08520.1 contains similarity... 49 8e-07
TC82245 similar to GP|16648720|gb|AAL25552.1 AT4g21620/F17L22_80... 48 1e-06
TC93504 weakly similar to PIR|S46286|S46286 RNA-binding protein ... 47 3e-06
BM814132 weakly similar to OMNI|MT3615.1 PE_PGRS family protein ... 46 5e-06
TC88887 weakly similar to GP|6523547|emb|CAB62280.1 hydroxyproli... 44 1e-05
>TC88280 weakly similar to GP|7024449|dbj|BAA92155.1 glycine-rich protein
{Citrus unshiu}, partial (56%)
Length = 603
Score = 288 bits (736), Expect = 6e-79
Identities = 133/133 (100%), Positives = 133/133 (100%)
Frame = +1
Query: 1 MDSKKAILILGLLAIILLISSEVSARDLTEASSNTQKEADDQTNELNDAKYGSYGGGYPG 60
MDSKKAILILGLLAIILLISSEVSARDLTEASSNTQKEADDQTNELNDAKYGSYGGGYPG
Sbjct: 37 MDSKKAILILGLLAIILLISSEVSARDLTEASSNTQKEADDQTNELNDAKYGSYGGGYPG 216
Query: 61 RGGGGNYGGGYPRNRGGYPGNRGGNYGGGYPGNRGGNYCRYGCCGDRYYGSCRKCCSYAG 120
RGGGGNYGGGYPRNRGGYPGNRGGNYGGGYPGNRGGNYCRYGCCGDRYYGSCRKCCSYAG
Sbjct: 217 RGGGGNYGGGYPRNRGGYPGNRGGNYGGGYPGNRGGNYCRYGCCGDRYYGSCRKCCSYAG 396
Query: 121 EAVAMQTENNTRN 133
EAVAMQTENNTRN
Sbjct: 397 EAVAMQTENNTRN 435
>TC76704 homologue to SP|Q09134|GRPA_MEDFA Abscisic acid and environmental
stress inducible protein. [Sickle medic] {Medicago
falcata}, partial (81%)
Length = 771
Score = 130 bits (328), Expect = 1e-31
Identities = 85/171 (49%), Positives = 101/171 (58%), Gaps = 38/171 (22%)
Frame = +2
Query: 1 MDSKKAILILGLLAIILLISSEVSARDLTEASSNTQKEADDQTNELNDAKYG-------- 52
MDSKKAILILGLLA++LLISSEVSARDLTE +S+ +KE ++TNE+NDAKYG
Sbjct: 38 MDSKKAILILGLLAMVLLISSEVSARDLTETTSDAKKEVVEKTNEVNDAKYGGGYNGGGY 217
Query: 53 SYGGGYPGRG---GGGNY--GGGYPRNRGGYPGNRGGNY---GGGYPG------------ 92
++GGGY G G GGG Y GGGY GGY N GG Y GGGY G
Sbjct: 218 NHGGGYNGGGYNHGGGGYNNGGGYNHGGGGY--NNGGGYNHGGGGYNGGGYNHGGGGYNG 391
Query: 93 ----------NRGGNYCRYGCCGDRYYGSCRKCCSYAGEAVAMQTENNTRN 133
N GG C+Y C G +CCS+A E VAMQ ++NT+N
Sbjct: 392 GGYNHGGGGYNHGGGGCQYHCHG--------RCCSHA-EFVAMQAKDNTQN 517
>AW690548 similar to SP|Q09134|GRPA Abscisic acid and environmental stress
inducible protein. [Sickle medic] {Medicago falcata},
partial (68%)
Length = 599
Score = 115 bits (288), Expect = 5e-27
Identities = 71/131 (54%), Positives = 82/131 (62%)
Frame = +1
Query: 1 MDSKKAILILGLLAIILLISSEVSARDLTEASSNTQKEADDQTNELNDAKYGSYGGGYPG 60
MDSKKAILILGLLA++LLISSEVSARDLTE SS+ +KE ++TNE+NDAKYG GGG
Sbjct: 22 MDSKKAILILGLLAMVLLISSEVSARDLTETSSDAKKEVVEKTNEVNDAKYG--GGGNYH 195
Query: 61 RGGGGNYGGGYPRNRGGYPGNRGGNYGGGYPGNRGGNYCRYGCCGDRYYGSCRKCCSYAG 120
GGG YG GG++ GG + GG C Y C G CCSYA
Sbjct: 196 NGGGHYYG--------------GGSHHGGGGSHHGGGGCHYYCHG--------HCCSYA- 306
Query: 121 EAVAMQTENNT 131
E VA+QTE T
Sbjct: 307 EFVAVQTEEKT 339
>TC76707 similar to SP|Q07202|CORA_MEDSA Cold and drought-regulated protein
CORA. [Alfalfa] {Medicago sativa}, partial (87%)
Length = 780
Score = 114 bits (284), Expect = 1e-26
Identities = 62/99 (62%), Positives = 73/99 (73%), Gaps = 1/99 (1%)
Frame = +1
Query: 1 MDSKKAILILGLLAIILLISSEVSARDLTEASSNTQKEADDQTNELNDAKYGSYGGGYPG 60
+DSKKAIL+LGLLA+ L ISS +SARDLTE S++T+KE ++TNE+NDAKYG YGGGY
Sbjct: 7 VDSKKAILMLGLLAMAL-ISSVMSARDLTETSTDTKKEVVEKTNEVNDAKYGGYGGGY-N 180
Query: 61 RGGGGNYGGGYPRNRGGYPGNRGG-NYGGGYPGNRGGNY 98
GGGG GGGY GGY GG N+GGG N GG Y
Sbjct: 181 HGGGGYNGGGYNHGGGGYNNGGGGYNHGGGGYNNGGGGY 297
Score = 55.5 bits (132), Expect = 6e-09
Identities = 28/67 (41%), Positives = 40/67 (58%)
Frame = +1
Query: 31 ASSNTQKEADDQTNELNDAKYGSYGGGYPGRGGGGNYGGGYPRNRGGYPGNRGGNYGGGY 90
A+ + + +++TNE+NDAKYG GGY GG N+GGG + GG + GG +GG
Sbjct: 469 AAESVAVQTEEKTNEVNDAKYG---GGYNHGGGSYNHGGGSYHHGGGGYNHGGGGHGGHG 639
Query: 91 PGNRGGN 97
G GG+
Sbjct: 640 GGGHGGH 660
>TC76705 similar to SP|Q07202|CORA_MEDSA Cold and drought-regulated protein
CORA. [Alfalfa] {Medicago sativa}, partial (80%)
Length = 854
Score = 111 bits (277), Expect = 9e-26
Identities = 76/144 (52%), Positives = 88/144 (60%), Gaps = 13/144 (9%)
Frame = +2
Query: 1 MDSKKAILILGLLAIILLISSEVSARDLTEASSNTQKEADDQTNELNDAKYGSY------ 54
MDSKKAIL+LGLLA+ L ISS +SARDLTE S++ +KE ++TNE+NDAKYG Y
Sbjct: 47 MDSKKAILMLGLLAMAL-ISSVMSARDLTETSTDAKKEVVEKTNEVNDAKYGGYNHGGYN 223
Query: 55 -GGGYPGRGGGGNYGGGYPRNRGGYPGNRGG--NYGGGYPGNRG----GNYCRYGCCGDR 107
GGGY GGG N+GGGY N GGY GG N GGGY G G G Y R G G R
Sbjct: 224 HGGGY--NGGGYNHGGGY-HNGGGYHNGGGGYHNGGGGYNGGGGHGGHGGYNRGGGHGGR 394
Query: 108 YYGSCRKCCSYAGEAVAMQTENNT 131
A E+VA+QTE T
Sbjct: 395 ----------GAAESVAVQTEEKT 436
Score = 48.1 bits (113), Expect = 1e-06
Identities = 25/66 (37%), Positives = 38/66 (56%), Gaps = 1/66 (1%)
Frame = +2
Query: 31 ASSNTQKEADDQTNELNDAKYGSYGGGYPGRGGGGNYG-GGYPRNRGGYPGNRGGNYGGG 89
A+ + + +++TNE+NDA+ G GG + G N+G G Y GGY + GG +GGG
Sbjct: 398 AAESVAVQTEEKTNEVNDARNGGGGGSFNKGGASYNHGRGSYHHG*GGY-NHGGGGHGGG 574
Query: 90 YPGNRG 95
G+ G
Sbjct: 575 GHGSHG 592
>TC76706 similar to SP|Q07202|CORA_MEDSA Cold and drought-regulated protein
CORA. [Alfalfa] {Medicago sativa}, partial (94%)
Length = 1034
Score = 108 bits (270), Expect = 6e-25
Identities = 74/144 (51%), Positives = 88/144 (60%), Gaps = 13/144 (9%)
Frame = +1
Query: 1 MDSKKAILILGLLAIILLISSEVSARDLTEASSNTQKEADDQTNELNDAKYGSY------ 54
MDSKKAIL+LGLLA+ L ISS +SAR+LTE S++ +KE ++TNE+NDAKYG Y
Sbjct: 37 MDSKKAILMLGLLAMAL-ISSVMSARELTETSTDAKKEVVEKTNEVNDAKYGGYNHGGYN 213
Query: 55 -GGGYPGRGGGGNYGGGYPRNRGGYPGNRGGNY---GGGYPGNRG-GNYCRYGCCGDR-- 107
GGGY GGG N+GGGY GGY N GG Y GGGY G G G + Y G
Sbjct: 214 HGGGY--NGGGYNHGGGYHNGGGGY-HNGGGGYNHGGGGYNGGGGHGGHGGYNGGGGHGG 384
Query: 108 YYGSCRKCCSYAGEAVAMQTENNT 131
Y G A E+VA+QTE T
Sbjct: 385 YNGGGGHGGHGAAESVAVQTEEKT 456
Score = 56.2 bits (134), Expect = 4e-09
Identities = 28/71 (39%), Positives = 42/71 (58%), Gaps = 4/71 (5%)
Frame = +1
Query: 31 ASSNTQKEADDQTNELNDAKYG----SYGGGYPGRGGGGNYGGGYPRNRGGYPGNRGGNY 86
A+ + + +++TNE+NDAKYG ++GG Y GG N+GGG + GG + GG +
Sbjct: 418 AAESVAVQTEEKTNEVNDAKYGGGSYNHGGSYNHGGGSYNHGGGSYHHGGGGYNHGGGGH 597
Query: 87 GGGYPGNRGGN 97
GG G GG+
Sbjct: 598 GGHGGGGHGGH 630
>TC89001 similar to GP|5726567|gb|AAD48471.1| glycine-rich RNA-binding
protein {Glycine max}, partial (90%)
Length = 488
Score = 57.4 bits (137), Expect = 2e-09
Identities = 42/97 (43%), Positives = 50/97 (51%), Gaps = 6/97 (6%)
Frame = +3
Query: 20 SSEVSARDLTEASSNTQKEADDQTNELNDAKYGSYGGGYPGRGGGGNYGGGYPRNRGGYP 79
+ E S RD E + ++ D + +N+A+ GGG G GGGG YGGG GGY
Sbjct: 198 AEEKSMRDAIEEMNG--QDIDGRNITVNEAQSRGSGGGGRG-GGGGGYGGG-----GGY- 350
Query: 80 GNRGGNYGGGYPGNRGGNYCR------YGCCGDRYYG 110
G GG YGGG G R G Y R YG GDR YG
Sbjct: 351 GGGGGGYGGG-GGRRDGGYSRSGGGGGYGGGGDRGYG 458
Score = 47.8 bits (112), Expect = 1e-06
Identities = 29/53 (54%), Positives = 29/53 (54%), Gaps = 5/53 (9%)
Frame = +3
Query: 51 YGSYGGGYPGRGGGGNYGGGYPRNRGGYP-GNRGGNYGG----GYPGNRGGNY 98
YG GGGY G GGG YGGG R GGY GG YGG GY G GG Y
Sbjct: 330 YGG-GGGYGG--GGGGYGGGGGRRDGGYSRSGGGGGYGGGGDRGYGGGGGGGY 479
Score = 45.1 bits (105), Expect = 8e-06
Identities = 22/38 (57%), Positives = 22/38 (57%)
Frame = +3
Query: 52 GSYGGGYPGRGGGGNYGGGYPRNRGGYPGNRGGNYGGG 89
G GGY GGGG YGGG R GY G GG YGGG
Sbjct: 384 GRRDGGYSRSGGGGGYGGGGDR---GYGGGGGGGYGGG 488
>TC86221 similar to PIR|T09608|T09608 environmental stress-induced protein -
alfalfa (fragment), partial (25%)
Length = 655
Score = 56.6 bits (135), Expect = 3e-09
Identities = 51/138 (36%), Positives = 69/138 (49%), Gaps = 5/138 (3%)
Frame = +1
Query: 1 MDSKKAILILGLLAIILLISSEVSARDLTEASSNTQKEADDQTNELNDAKYGSY-----G 55
M SKKAIL+LGLLA++ LISS +S E S++ ++ +QT +LN K G Y
Sbjct: 43 MVSKKAILMLGLLAMV-LISSVIS-----ETSTDPNWDSVEQT-DLNYGKDGGYVGGSNN 201
Query: 56 GGYPGRGGGGNYGGGYPRNRGGYPGNRGGNYGGGYPGNRGGNYCRYGCCGDRYYGSCRKC 115
GG G N G G +GG+ N GG GY G GG+ +G G +G
Sbjct: 202 GGDSNNGDDSNNGDGDVHEKGGHVYNHGGE---GYNGGGGGH--NHGRRGHGGHG----- 351
Query: 116 CSYAGEAVAMQTENNTRN 133
A++TE+ TRN
Sbjct: 352 --------AVETEDETRN 381
>TC84888 similar to GP|21322752|dbj|BAB78536. cold shock protein-1 {Triticum
aestivum}, partial (58%)
Length = 612
Score = 54.7 bits (130), Expect = 1e-08
Identities = 33/75 (44%), Positives = 41/75 (54%), Gaps = 6/75 (8%)
Frame = +2
Query: 28 LTEASSNTQKEADDQTNELNDAKYGS------YGGGYPGRGGGGNYGGGYPRNRGGYPGN 81
+ EA + + +A D T G+ YGGG GRGGGG YGGGY + GG G+
Sbjct: 206 IVEAGDDGRTKAIDVTGPNGAPPQGAPRQEMGYGGG-GGRGGGG-YGGGYDQGYGGGGGS 379
Query: 82 RGGNYGGGYPGNRGG 96
GG +GGG G RGG
Sbjct: 380 YGGGFGGGRGGGRGG 424
Score = 52.0 bits (123), Expect = 7e-08
Identities = 32/62 (51%), Positives = 35/62 (55%), Gaps = 3/62 (4%)
Frame = +2
Query: 52 GSYGGGYPG--RGGGGNYGGGYPRNRGGYPGNRGGNYG-GGYPGNRGGNYCRYGCCGDRY 108
G YGGGY GGGG+YGGG+ RGG G GG Y GGY G GG Y + G G
Sbjct: 329 GGYGGGYDQGYGGGGGSYGGGFGGGRGG--GRGGGGYXQGGYGG--GGGYXQGGYGGQGG 496
Query: 109 YG 110
YG
Sbjct: 497 YG 502
Score = 47.8 bits (112), Expect = 1e-06
Identities = 26/48 (54%), Positives = 28/48 (58%), Gaps = 1/48 (2%)
Frame = +2
Query: 52 GSYGGGYPGRGGGGNYGGGYPRNRGGYPGNRGGNYGG-GYPGNRGGNY 98
GSYGGG+ G GGG GGGY +GGY G G GG G G GG Y
Sbjct: 374 GSYGGGFGGGRGGGRGGGGY--XQGGYGGGGGYXQGGYGGQGGYGGGY 511
>TC79557 similar to GP|12039289|gb|AAG46079.1 hypothetical protein {Oryza
sativa}, partial (45%)
Length = 754
Score = 53.9 bits (128), Expect = 2e-08
Identities = 35/94 (37%), Positives = 49/94 (51%), Gaps = 1/94 (1%)
Frame = +1
Query: 5 KAILILGLLAIILLISSEVSARDLTEASSNTQKEADDQTNELNDAKYGSYGGGYPGRGGG 64
K I IL L I+L +++ VS+R T +++ AD+ N + G+ GG G G G
Sbjct: 49 KQISILILFTILLTVTTSVSSRRST-STTKPSPSADNNNNNKDGVGGGNDYGGSFGFGPG 225
Query: 65 GNYG-GGYPRNRGGYPGNRGGNYGGGYPGNRGGN 97
G + G+ N G GN G YGGGY G GG+
Sbjct: 226 GGFSIPGFDNNIIGGGGNYAGGYGGGYGGPNGGH 327
>TC76557 similar to PIR|S71779|S71779 glycine-rich RNA-binding protein GRP1
- wheat, partial (95%)
Length = 912
Score = 52.4 bits (124), Expect = 5e-08
Identities = 33/80 (41%), Positives = 41/80 (51%), Gaps = 1/80 (1%)
Frame = +1
Query: 20 SSEVSARDLTEASSNTQKEADDQTNELNDAKYGSYGGGYPGRGGGGNYGGGYPRNRGGYP 79
++E S D EA + ++ D + +N A+ GGG GGGG GGGY GGY
Sbjct: 406 ANEKSMNDAIEAMNG--QDLDGRNITVNQAQSRGSGGG----GGGGRGGGGYGGGGGGYG 567
Query: 80 GNRGG-NYGGGYPGNRGGNY 98
G R G GGGY G GG Y
Sbjct: 568 GERRGYGGGGGYGGGGGGGY 627
Score = 50.1 bits (118), Expect = 3e-07
Identities = 37/104 (35%), Positives = 46/104 (43%), Gaps = 8/104 (7%)
Frame = +1
Query: 14 AIILLISSEVSARDLTEASSNTQKEADDQTNELNDAKYGSYGGGYPGR----GGGGNYGG 69
AI + ++ R++T + ++ YG GGGY G GGGG YGG
Sbjct: 430 AIEAMNGQDLDGRNITVNQAQSRGSGGGGGGGRGGGGYGGGGGGYGGERRGYGGGGGYGG 609
Query: 70 GYPRNRGGYPGNRGG----NYGGGYPGNRGGNYCRYGCCGDRYY 109
G GGY RGG + GGG G GG Y R G GD Y
Sbjct: 610 G---GGGGYGERRGGGGGYSRGGGGGGYGGGGYSRGG--GDGGY 726
>TC76561 similar to PIR|S71779|S71779 glycine-rich RNA-binding protein GRP1
- wheat, partial (95%)
Length = 1054
Score = 52.4 bits (124), Expect = 5e-08
Identities = 33/80 (41%), Positives = 41/80 (51%), Gaps = 1/80 (1%)
Frame = +1
Query: 20 SSEVSARDLTEASSNTQKEADDQTNELNDAKYGSYGGGYPGRGGGGNYGGGYPRNRGGYP 79
++E S D EA + ++ D + +N A+ GGG GGGG GGGY GGY
Sbjct: 502 ANEKSMNDAIEAMNG--QDLDGRNITVNQAQSRGSGGG----GGGGRGGGGYGGGGGGYG 663
Query: 80 GNRGG-NYGGGYPGNRGGNY 98
G R G GGGY G GG Y
Sbjct: 664 GERRGYGGGGGYGGGGGGGY 723
Score = 50.1 bits (118), Expect = 3e-07
Identities = 37/104 (35%), Positives = 46/104 (43%), Gaps = 8/104 (7%)
Frame = +1
Query: 14 AIILLISSEVSARDLTEASSNTQKEADDQTNELNDAKYGSYGGGYPGR----GGGGNYGG 69
AI + ++ R++T + ++ YG GGGY G GGGG YGG
Sbjct: 526 AIEAMNGQDLDGRNITVNQAQSRGSGGGGGGGRGGGGYGGGGGGYGGERRGYGGGGGYGG 705
Query: 70 GYPRNRGGYPGNRGG----NYGGGYPGNRGGNYCRYGCCGDRYY 109
G GGY RGG + GGG G GG Y R G GD Y
Sbjct: 706 G---GGGGYGERRGGGGGYSRGGGGGGYGGGGYSRGG--GDGGY 822
>TC76556 similar to PIR|S71779|S71779 glycine-rich RNA-binding protein GRP1
- wheat, partial (95%)
Length = 754
Score = 52.4 bits (124), Expect = 5e-08
Identities = 33/80 (41%), Positives = 41/80 (51%), Gaps = 1/80 (1%)
Frame = +2
Query: 20 SSEVSARDLTEASSNTQKEADDQTNELNDAKYGSYGGGYPGRGGGGNYGGGYPRNRGGYP 79
++E S D EA + ++ D + +N A+ GGG GGGG GGGY GGY
Sbjct: 248 ANEKSMNDAIEAMNG--QDLDGRNITVNQAQSRGSGGG----GGGGRGGGGYGGGGGGYG 409
Query: 80 GNRGG-NYGGGYPGNRGGNY 98
G R G GGGY G GG Y
Sbjct: 410 GERRGYGGGGGYGGGGGGGY 469
Score = 50.1 bits (118), Expect = 3e-07
Identities = 37/104 (35%), Positives = 46/104 (43%), Gaps = 8/104 (7%)
Frame = +2
Query: 14 AIILLISSEVSARDLTEASSNTQKEADDQTNELNDAKYGSYGGGYPGR----GGGGNYGG 69
AI + ++ R++T + ++ YG GGGY G GGGG YGG
Sbjct: 272 AIEAMNGQDLDGRNITVNQAQSRGSGGGGGGGRGGGGYGGGGGGYGGERRGYGGGGGYGG 451
Query: 70 GYPRNRGGYPGNRGG----NYGGGYPGNRGGNYCRYGCCGDRYY 109
G GGY RGG + GGG G GG Y R G GD Y
Sbjct: 452 G---GGGGYGERRGGGGGYSRGGGGGGYGGGGYSRGG--GDGGY 568
>TC87796 weakly similar to SP|P03211|EBN1_EBV EBNA-1 nuclear protein.
[strain B95-8 Human herpesvirus 4] {Epstein-barr
virus}, partial (23%)
Length = 431
Score = 51.2 bits (121), Expect = 1e-07
Identities = 26/50 (52%), Positives = 27/50 (54%)
Frame = +1
Query: 52 GSYGGGYPGRGGGGNYGGGYPRNRGGYPGNRGGNYGGGYPGNRGGNYCRY 101
G YGGG PG GGGG GG GG G GG YGGG G GG + Y
Sbjct: 112 GGYGGGSPGGGGGGGGSGG----GGGGGGAHGGAYGGGIGGGEGGGHGGY 249
Score = 46.2 bits (108), Expect = 4e-06
Identities = 23/45 (51%), Positives = 23/45 (51%)
Frame = +1
Query: 52 GSYGGGYPGRGGGGNYGGGYPRNRGGYPGNRGGNYGGGYPGNRGG 96
G YGGG GGGG GG G G GG YGGG PG GG
Sbjct: 16 GGYGGGGGSGGGGGGGAGGAHGVGYGSGGGTGGGYGGGSPGGGGG 150
Score = 28.9 bits (63), Expect = 0.62
Identities = 18/46 (39%), Positives = 20/46 (43%)
Frame = +1
Query: 60 GRGGGGNYGGGYPRNRGGYPGNRGGNYGGGYPGNRGGNYCRYGCCG 105
GR GG YGGG GG+ GGG G G + YG G
Sbjct: 1 GRDHGGGYGGG------------GGSGGGGGGGAGGAHGVGYGSGG 102
>AL372330 similar to GP|21322752|dbj cold shock protein-1 {Triticum
aestivum}, partial (33%)
Length = 435
Score = 50.8 bits (120), Expect = 2e-07
Identities = 27/47 (57%), Positives = 28/47 (59%)
Frame = +2
Query: 52 GSYGGGYPGRGGGGNYGGGYPRNRGGYPGNRGGNYGGGYPGNRGGNY 98
G YGGG G GGG YGGG + GGY G GG GGGY G GG Y
Sbjct: 179 GGYGGGASGGYGGGGYGGG---SGGGYGG--GGYGGGGYGGGGGGGY 304
Score = 50.4 bits (119), Expect = 2e-07
Identities = 26/55 (47%), Positives = 28/55 (50%), Gaps = 8/55 (14%)
Frame = +2
Query: 52 GSYGGG----YPGRGGGGNYGGG----YPRNRGGYPGNRGGNYGGGYPGNRGGNY 98
G YGG YP GG YGGG Y + GGY G+ GG GGGY G G Y
Sbjct: 47 GGYGGSSSSSYPASSSGGGYGGGSSGGYGGSSGGYGGSSGGYGGGGYGGGASGGY 211
Score = 48.9 bits (115), Expect = 6e-07
Identities = 25/46 (54%), Positives = 25/46 (54%)
Frame = +2
Query: 52 GSYGGGYPGRGGGGNYGGGYPRNRGGYPGNRGGNYGGGYPGNRGGN 97
G YGGG G G GG YGGG GGY G GG YGG G G N
Sbjct: 203 GGYGGGGYGGGSGGGYGGG-GYGGGGYGGGGGGGYGGDKMGALGAN 337
Score = 48.1 bits (113), Expect = 1e-06
Identities = 32/65 (49%), Positives = 32/65 (49%), Gaps = 6/65 (9%)
Frame = +2
Query: 52 GSYGGGYPGRGGG--GNYGG---GYPRNRGGYPGNRGGNY-GGGYPGNRGGNYCRYGCCG 105
G YGGG G GG G YGG GY GGY G G Y GGGY G GG Y G G
Sbjct: 98 GGYGGGSSGGYGGSSGGYGGSSGGY--GGGGYGGGASGGYGGGGYGGGSGGGYGGGGYGG 271
Query: 106 DRYYG 110
Y G
Sbjct: 272 GGYGG 286
Score = 43.9 bits (102), Expect = 2e-05
Identities = 27/58 (46%), Positives = 28/58 (47%), Gaps = 1/58 (1%)
Frame = +2
Query: 51 YGSYGGGYPGRGGGGNYGGGYPRNRGGYPGNRGGN-YGGGYPGNRGGNYCRYGCCGDR 107
YG GGY G G GG GGY GGY G GG GGGY G G G GD+
Sbjct: 149 YGGSSGGYGGGGYGGGASGGY--GGGGYGGGSGGGYGGGGYGGGGYGGGGGGGYGGDK 316
Score = 42.4 bits (98), Expect = 5e-05
Identities = 27/67 (40%), Positives = 29/67 (42%), Gaps = 2/67 (2%)
Frame = +2
Query: 32 SSNTQKEADDQTNELNDAKYGSYGGGYPGRGG--GGNYGGGYPRNRGGYPGNRGGNYGGG 89
SS++ A G YGG G GG GG GGGY G G GG YGGG
Sbjct: 62 SSSSSYPASSSGGGYGGGSSGGYGGSSGGYGGSSGGYGGGGYGGGASG--GYGGGGYGGG 235
Query: 90 YPGNRGG 96
G GG
Sbjct: 236 SGGGYGG 256
Score = 40.0 bits (92), Expect = 3e-04
Identities = 23/50 (46%), Positives = 26/50 (52%), Gaps = 5/50 (10%)
Frame = +2
Query: 52 GSYGGGYPGRGGGGNYGGGYPRNR--GGYPGNRGGNYG---GGYPGNRGG 96
G YGG G G GG+ YP + GGY G G YG GGY G+ GG
Sbjct: 23 GGYGGSSSG-GYGGSSSSSYPASSSGGGYGGGSSGGYGGSSGGYGGSSGG 169
Score = 39.7 bits (91), Expect = 4e-04
Identities = 25/59 (42%), Positives = 28/59 (47%), Gaps = 1/59 (1%)
Frame = +2
Query: 53 SYGGGYPGRGGGGNYGGGYPRNRGGYPGNR-GGNYGGGYPGNRGGNYCRYGCCGDRYYG 110
S GGY G GG YGG + YP + GG YGGG G GG+ YG Y G
Sbjct: 14 SSSGGYGGSSSGG-YGGS---SSSSYPASSSGGGYGGGSSGGYGGSSGGYGGSSGGYGG 178
>TC86535 similar to GP|9758076|dbj|BAB08520.1 contains similarity to RNA
binding protein~gene_id:MNF13.1 {Arabidopsis thaliana},
partial (48%)
Length = 1555
Score = 48.5 bits (114), Expect = 8e-07
Identities = 33/86 (38%), Positives = 40/86 (46%), Gaps = 14/86 (16%)
Frame = +1
Query: 27 DLTEASSNTQKEADDQTNELNDAK--YGS--YGGGYPGRGGG-GNYGGGYPRNRGGYPGN 81
++ +A + ND++ YGS YG Y G GGG G GGGY R+ Y G
Sbjct: 748 EIKKAEPKKANAPPPSSKRYNDSRSSYGSGGYGDAYDGFGGGFGGVGGGYSRSGSAY-GG 924
Query: 82 RGGNYG---------GGYPGNRGGNY 98
RGG YG GGY G GG Y
Sbjct: 925 RGGPYGGYGSEFAGYGGYAGAMGGPY 1002
Score = 41.6 bits (96), Expect = 9e-05
Identities = 25/64 (39%), Positives = 31/64 (48%), Gaps = 5/64 (7%)
Frame = +1
Query: 51 YGSYGGGYPGRGGGGNYGGGYPRNRGGYPGNRGGNYGG-----GYPGNRGGNYCRYGCCG 105
YG+YGG G GG G + GY + GG YGG + G+RGG YG G
Sbjct: 1096 YGAYGGAGAGAAAGGGGSSGAGAYQSGYDASLGGGYGGAASGASFYGSRGG----YGGAG 1263
Query: 106 DRYY 109
RY+
Sbjct: 1264 -RYH 1272
Score = 33.1 bits (74), Expect = 0.033
Identities = 21/63 (33%), Positives = 25/63 (39%)
Frame = +1
Query: 49 AKYGSYGGGYPGRGGGGNYGGGYPRNRGGYPGNRGGNYGGGYPGNRGGNYCRYGCCGDRY 108
+ Y G G PG G G G GG + G Y GY + GG Y G +
Sbjct: 1054 SSYDLSGYGGPGESYGAYGGAGAGAAAGGGGSSGAGAYQSGYDASLGGGY-GGAASGASF 1230
Query: 109 YGS 111
YGS
Sbjct: 1231 YGS 1239
Score = 31.2 bits (69), Expect = 0.12
Identities = 24/67 (35%), Positives = 25/67 (36%), Gaps = 21/67 (31%)
Frame = +1
Query: 51 YGSYG------GGYPGRGGG-----------GNYGGG----YPRNRGGYPGNRGGNYGGG 89
YG YG GGY G GG G YGG Y + G PG G YGG
Sbjct: 937 YGGYGSEFAGYGGYAGAMGGPYRGDPSLAYAGRYGGSFRSSYDLSGYGGPGESYGAYGGA 1116
Query: 90 YPGNRGG 96
G G
Sbjct: 1117 GAGAAAG 1137
>TC82245 similar to GP|16648720|gb|AAL25552.1 AT4g21620/F17L22_80
{Arabidopsis thaliana}, partial (67%)
Length = 604
Score = 47.8 bits (112), Expect = 1e-06
Identities = 36/102 (35%), Positives = 51/102 (49%), Gaps = 8/102 (7%)
Frame = +3
Query: 4 KKAILILGLLAIILLISSEVSARDLTEASS-NTQ-KEADDQTNELNDAKYGSYGGGYPGR 61
K L L ++L+++S ++ R ++ SS +TQ K D+ N G GG PG
Sbjct: 78 KTMTTFLLLTTLLLIVTSSLATRPVSSGSSPSTQVKHPKDKNNN-----QGGGGGSIPGV 242
Query: 62 GG--GGNYG--GGY--PRNRGGYPGNRGGNYGGGYPGNRGGN 97
GG GG +G GG+ P G+ GG YG GY G GG+
Sbjct: 243 GGIPGGYFGPGGGFNIPGFGNGFGNGVGGGYGSGYGGPNGGH 368
>TC93504 weakly similar to PIR|S46286|S46286 RNA-binding protein - wood
tobacco, partial (55%)
Length = 576
Score = 46.6 bits (109), Expect = 3e-06
Identities = 37/77 (48%), Positives = 40/77 (51%), Gaps = 8/77 (10%)
Frame = +3
Query: 30 EASSNTQ----KEADDQTNELN----DAKYGSYGGGYPGRGGGGNYGGGYPRNRGGYPGN 81
EASS Q KE D +T +N A+ G GGGY GGGG YGGG GGY G
Sbjct: 300 EASSALQAMDNKELDGRTVRVNYATERARPGFGGGGY---GGGGGYGGG-----GGYGG- 452
Query: 82 RGGNYGGGYPGNRGGNY 98
GG Y GG GGNY
Sbjct: 453 -GGGYSGG-----GGNY 485
Score = 32.7 bits (73), Expect = 0.043
Identities = 18/41 (43%), Positives = 21/41 (50%), Gaps = 2/41 (4%)
Frame = +3
Query: 52 GSYGGGYPGRGGGGNYGGGYPRN--RGGYPGNRGGNYGGGY 90
G YGGG GGGGNYGG + + R + G GG Y
Sbjct: 438 GGYGGGGGYSGGGGNYGGWWRKLWWRKLWVAETTGGVGGRY 560
>BM814132 weakly similar to OMNI|MT3615.1 PE_PGRS family protein
{Mycobacterium tuberculosis CDC1551}, partial (7%)
Length = 164
Score = 45.8 bits (107), Expect = 5e-06
Identities = 24/45 (53%), Positives = 25/45 (55%)
Frame = +2
Query: 52 GSYGGGYPGRGGGGNYGGGYPRNRGGYPGNRGGNYGGGYPGNRGG 96
G+ GGG G GGGG GGG GG G RGG GGG G GG
Sbjct: 8 GAGGGGGGGGGGGGGGGGGGGGGGGGGGGGRGGGGGGGGGGGGGG 142
Score = 45.4 bits (106), Expect = 6e-06
Identities = 24/45 (53%), Positives = 24/45 (53%)
Frame = +2
Query: 52 GSYGGGYPGRGGGGNYGGGYPRNRGGYPGNRGGNYGGGYPGNRGG 96
G GGG G GGGG GGG RGG G GG GGG G GG
Sbjct: 29 GGGGGGGGGGGGGGGGGGGGGGGRGGGGGGGGGGGGGGGGGGGGG 163
Score = 44.7 bits (104), Expect = 1e-05
Identities = 23/42 (54%), Positives = 23/42 (54%)
Frame = +1
Query: 55 GGGYPGRGGGGNYGGGYPRNRGGYPGNRGGNYGGGYPGNRGG 96
GGG G GGGG GGG RGG G GG GGG G GG
Sbjct: 4 GGGGGGGGGGGGGGGGGGGGRGGGGGGGGGGAGGGGGGGGGG 129
Score = 43.5 bits (101), Expect = 2e-05
Identities = 23/45 (51%), Positives = 23/45 (51%)
Frame = +1
Query: 52 GSYGGGYPGRGGGGNYGGGYPRNRGGYPGNRGGNYGGGYPGNRGG 96
G GGG G GGGG GGG GG G GG GGG G GG
Sbjct: 19 GGGGGGGGGGGGGGGRGGGGGGGGGGAGGGGGGGGGGGGGGGGGG 153
Score = 43.5 bits (101), Expect = 2e-05
Identities = 23/45 (51%), Positives = 23/45 (51%)
Frame = +3
Query: 52 GSYGGGYPGRGGGGNYGGGYPRNRGGYPGNRGGNYGGGYPGNRGG 96
G GGG G GGGG GGG GG G GG GGG G GG
Sbjct: 6 GGRGGGGGGGGGGGGGGGGAGGGGGGGGGGGGGGGGGGGGGGGGG 140
Score = 43.1 bits (100), Expect = 3e-05
Identities = 23/45 (51%), Positives = 23/45 (51%)
Frame = +3
Query: 52 GSYGGGYPGRGGGGNYGGGYPRNRGGYPGNRGGNYGGGYPGNRGG 96
G GGG G GGGG GGG GG G GG GGG G GG
Sbjct: 18 GGGGGGGGGGGGGGGAGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 152
Score = 43.1 bits (100), Expect = 3e-05
Identities = 23/45 (51%), Positives = 23/45 (51%)
Frame = +1
Query: 52 GSYGGGYPGRGGGGNYGGGYPRNRGGYPGNRGGNYGGGYPGNRGG 96
G GGG G GGGG GGG R GG G G GGG G GG
Sbjct: 1 GGGGGGGGGGGGGGGGGGGGGRGGGGGGGGGGAGGGGGGGGGGGG 135
Score = 40.4 bits (93), Expect = 2e-04
Identities = 22/45 (48%), Positives = 22/45 (48%)
Frame = +3
Query: 52 GSYGGGYPGRGGGGNYGGGYPRNRGGYPGNRGGNYGGGYPGNRGG 96
G GGG G GGG GGG GG G GG GGG G GG
Sbjct: 24 GGGGGGGGGGGGGAGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 158
Score = 40.4 bits (93), Expect = 2e-04
Identities = 22/45 (48%), Positives = 22/45 (48%)
Frame = +3
Query: 52 GSYGGGYPGRGGGGNYGGGYPRNRGGYPGNRGGNYGGGYPGNRGG 96
G GGG G GG G GGG GG G GG GGG G GG
Sbjct: 27 GGGGGGGGGGGGAGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 161
Score = 40.4 bits (93), Expect = 2e-04
Identities = 22/45 (48%), Positives = 22/45 (48%)
Frame = +3
Query: 52 GSYGGGYPGRGGGGNYGGGYPRNRGGYPGNRGGNYGGGYPGNRGG 96
G GGG G GGGG GG GG G GG GGG G GG
Sbjct: 15 GGGGGGGGGGGGGGGGAGGGGGGGGGGGGGGGGGGGGGGGGGGGG 149
Score = 30.8 bits (68), Expect = 0.16
Identities = 20/47 (42%), Positives = 20/47 (42%)
Frame = +2
Query: 56 GGYPGRGGGGNYGGGYPRNRGGYPGNRGGNYGGGYPGNRGGNYCRYG 102
GG G GGGG GGG GG GGG G GG R G
Sbjct: 2 GGGAGGGGGGGGGGG------------GGGGGGGGGGGGGGGGGRGG 106
>TC88887 weakly similar to GP|6523547|emb|CAB62280.1 hydroxyproline-rich
glycoprotein DZ-HRGP {Volvox carteri f. nagariensis},
partial (18%)
Length = 1681
Score = 44.3 bits (103), Expect = 1e-05
Identities = 27/58 (46%), Positives = 35/58 (59%), Gaps = 6/58 (10%)
Frame = -3
Query: 45 ELNDAKYGSYGG--GYPGRGG----GGNYGGGYPRNRGGYPGNRGGNYGGGYPGNRGG 96
E+ + ++ GG G+ GRGG G +GGGY RGG+ G RGG +GGGY RGG
Sbjct: 755 EVREDRFAGAGGPMGFGGRGGFGGMRGGFGGGY--GRGGFNGGRGG-FGGGY--GRGG 597
Score = 36.2 bits (82), Expect = 0.004
Identities = 23/47 (48%), Positives = 27/47 (56%), Gaps = 2/47 (4%)
Frame = -3
Query: 52 GSYGGGYPGRGGGGNYG-GGYPRNRGGYPGNRGGNYG-GGYPGNRGG 96
G +GG G GGG YG GG+ RGG+ GG YG GG+ G GG
Sbjct: 698 GGFGGMRGGFGGG--YGRGGFNGGRGGF----GGGYGRGGFAGGGGG 576
Score = 34.7 bits (78), Expect = 0.011
Identities = 18/41 (43%), Positives = 22/41 (52%)
Frame = -2
Query: 49 AKYGSYGGGYPGRGGGGNYGGGYPRNRGGYPGNRGGNYGGG 89
A G +GGG G GG G GG +P GG+ G G+Y G
Sbjct: 1098 ANRGGFGGGPGGPGGPGGPGGMHP---GGFGGGGPGSYSTG 985
Score = 26.9 bits (58), Expect = 2.4
Identities = 17/44 (38%), Positives = 18/44 (40%), Gaps = 6/44 (13%)
Frame = -2
Query: 70 GYPRNRGGYPGNRGGNYGGGYPGNR------GGNYCRYGCCGDR 107
G NRGG+ G GG G G PG GG Y G R
Sbjct: 1107 GATANRGGFGGGPGGPGGPGGPGGMHPGGFGGGGPGSYSTGGSR 976
Score = 26.9 bits (58), Expect = 2.4
Identities = 16/43 (37%), Positives = 19/43 (43%)
Frame = -2
Query: 60 GRGGGGNYGGGYPRNRGGYPGNRGGNYGGGYPGNRGGNYCRYG 102
G GGG GG PG GG + GG+ G G+Y G
Sbjct: 1086 GFGGGPGGPGG--------PGGPGGMHPGGFGGGGPGSYSTGG 982
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.313 0.139 0.433
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,288,629
Number of Sequences: 36976
Number of extensions: 78169
Number of successful extensions: 3706
Number of sequences better than 10.0: 355
Number of HSP's better than 10.0 without gapping: 1106
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2017
length of query: 133
length of database: 9,014,727
effective HSP length: 86
effective length of query: 47
effective length of database: 5,834,791
effective search space: 274235177
effective search space used: 274235177
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 53 (25.0 bits)
Medicago: description of AC138451.12


