

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
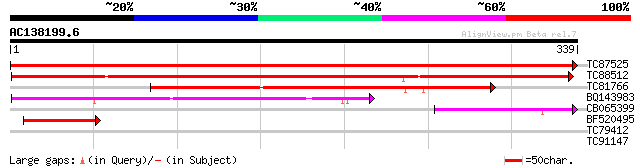
Query= AC138199.6 + phase: 0
(339 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC87525 similar to GP|9294498|dbj|BAB02717.1 aldose 1-epimerase-... 699 0.0
TC88512 weakly similar to PIR|T01933|T01933 probable aldose 1-ep... 344 2e-95
TC81766 weakly similar to GP|15824565|gb|AAL09397.1 plasmodesmal... 189 1e-48
BQ143983 74 6e-14
CB065399 OMNI|NTL01EC00 galactose-1-epimerase (mutarotase) {Esch... 57 8e-09
BF520495 weakly similar to PIR|T01933|T019 probable aldose 1-epi... 47 1e-05
TC79412 similar to GP|15809663|gb|AAL07275.1 COP9 complex subuni... 30 1.0
TC91147 28 5.1
>TC87525 similar to GP|9294498|dbj|BAB02717.1 aldose 1-epimerase-like
protein {Arabidopsis thaliana}, partial (94%)
Length = 1331
Score = 699 bits (1804), Expect = 0.0
Identities = 339/339 (100%), Positives = 339/339 (100%)
Frame = +3
Query: 1 MADQNKKPEIFELNNGTMQLLVTNLGCTITSFSVPAKDGVLSDVVLGLDSVESYQKGLAP 60
MADQNKKPEIFELNNGTMQLLVTNLGCTITSFSVPAKDGVLSDVVLGLDSVESYQKGLAP
Sbjct: 36 MADQNKKPEIFELNNGTMQLLVTNLGCTITSFSVPAKDGVLSDVVLGLDSVESYQKGLAP 215
Query: 61 YFGCIVGRVANRIKNGKFTLDGVEYSLPLNKAPNTLHGGNVGFDKKVWDVLEYKKGETPS 120
YFGCIVGRVANRIKNGKFTLDGVEYSLPLNKAPNTLHGGNVGFDKKVWDVLEYKKGETPS
Sbjct: 216 YFGCIVGRVANRIKNGKFTLDGVEYSLPLNKAPNTLHGGNVGFDKKVWDVLEYKKGETPS 395
Query: 121 ITFKYDSHDGEEGYPGDITVTATYTLTSSTTMRLDMEGVAKNKPTIINLAQHTYWNLAGH 180
ITFKYDSHDGEEGYPGDITVTATYTLTSSTTMRLDMEGVAKNKPTIINLAQHTYWNLAGH
Sbjct: 396 ITFKYDSHDGEEGYPGDITVTATYTLTSSTTMRLDMEGVAKNKPTIINLAQHTYWNLAGH 575
Query: 181 SSGNILDHSIKISANHVTPVDQNTVPTGEIVPVKGTPFDFTSEKRIGDTINQVGLGYDHN 240
SSGNILDHSIKISANHVTPVDQNTVPTGEIVPVKGTPFDFTSEKRIGDTINQVGLGYDHN
Sbjct: 576 SSGNILDHSIKISANHVTPVDQNTVPTGEIVPVKGTPFDFTSEKRIGDTINQVGLGYDHN 755
Query: 241 YVLDCGEEKAGLRHAAKVRDPSSSRVLNLWTNAPGVQFYTGNYVDNVTGKGGAVYGKHAG 300
YVLDCGEEKAGLRHAAKVRDPSSSRVLNLWTNAPGVQFYTGNYVDNVTGKGGAVYGKHAG
Sbjct: 756 YVLDCGEEKAGLRHAAKVRDPSSSRVLNLWTNAPGVQFYTGNYVDNVTGKGGAVYGKHAG 935
Query: 301 LCLETQGFPDAVNKPNFPSVVVKPGEKYQHSMLFEFSIE 339
LCLETQGFPDAVNKPNFPSVVVKPGEKYQHSMLFEFSIE
Sbjct: 936 LCLETQGFPDAVNKPNFPSVVVKPGEKYQHSMLFEFSIE 1052
>TC88512 weakly similar to PIR|T01933|T01933 probable aldose 1-epimerase (EC
5.1.3.3) - common tobacco, partial (76%)
Length = 1323
Score = 344 bits (883), Expect = 2e-95
Identities = 174/339 (51%), Positives = 226/339 (66%), Gaps = 3/339 (0%)
Frame = +3
Query: 2 ADQNKKPEIFELNNGTMQLLVTNLGCTITSFSVPAKDGVLSDVVLGLDSVESYQKGLAPY 61
+D+ +K IFEL G + L VTN G +I S +P K+G L+D+VLG DS++ Y + Y
Sbjct: 180 SDEKEKVGIFELKKGGLSLKVTNWGASIVSLVLPDKNGKLADIVLGYDSIKEYTND-STY 356
Query: 62 FGCIVGRVANRIKNGKFTLDGVEYSLPLNKAPNTLHGGNVGFDKKVWDVLEYKK-GETPS 120
FG VGRVANRI +F L+G Y L N+ NTLHGG+ GF +W V Y+K G +PS
Sbjct: 357 FGATVGRVANRIGGAQFNLNGKHYKLVANEGNNTLHGGSRGFSDVIWKVKRYQKEGPSPS 536
Query: 121 ITFKYDSHDGEEGYPGDITVTATYTLTSSTTMRLDMEGVAKNKPTIINLAQHTYWNLAGH 180
+TF Y S DGEEG+PGD+ T +Y LT + + M+ A NKPT +NLA H YWN+ G
Sbjct: 537 VTFTYHSFDGEEGFPGDLLATVSYILTGKNQLVIIMKAKALNKPTPVNLANHAYWNIGGQ 716
Query: 181 SSGNILDHSIKISANHVTPVDQNTVPTGEIVPVKGTPFDFTSEKRIGDTINQVG--LGYD 238
+SGNIL+ I+I + T VD +PTG+ VKGT +DF + IG I+Q+ GYD
Sbjct: 717 NSGNILNEVIQIFGSKTTIVDDKLIPTGKFASVKGTAYDFLKPQIIGSRISQLAQHKGYD 896
Query: 239 HNYVLDCGEEKAGLRHAAKVRDPSSSRVLNLWTNAPGVQFYTGNYVDNVTGKGGAVYGKH 298
NYVLD GE+ ++ AAKV D S R+L L+TNAPG+QFYTGNY+ +V GK G VY H
Sbjct: 897 INYVLD-GEKGKKIKLAAKVHDKKSGRILELYTNAPGLQFYTGNYIKDVKGKSGYVYKAH 1073
Query: 299 AGLCLETQGFPDAVNKPNFPSVVVKPGEKYQHSMLFEFS 337
AGLCLE+Q FPD+VN PNFPS +V P + Y+H MLF+FS
Sbjct: 1074AGLCLESQAFPDSVNHPNFPSTIVTPEKPYKHYMLFKFS 1190
>TC81766 weakly similar to GP|15824565|gb|AAL09397.1 plasmodesmal receptor
{Nicotiana tabacum}, partial (48%)
Length = 635
Score = 189 bits (481), Expect = 1e-48
Identities = 104/211 (49%), Positives = 132/211 (62%), Gaps = 5/211 (2%)
Frame = +2
Query: 85 YSLPLNKAPNTLHGGNVGFDKKVWDVLEY-KKGETPSITFKYDSHDGEEGYPGDITVTAT 143
Y L N+ NTLHGG GF +W V +Y ++G+ P I F Y S DGEEG+PGD+ VT +
Sbjct: 5 YKLIANEGNNTLHGGPRGFSDVLWKVEKYVREGDQPLIKFFYHSFDGEEGFPGDLKVTVS 184
Query: 144 YTLTSSTTMRLDMEGVAKNKPTIINLAQHTYWNLAGHSSGNILDHSIKISANHVTPVDQN 203
Y L ++ + + M+ A NKPT INL H YWNL H+SGNILD ++I + TP D N
Sbjct: 185 YILRKNS-LTIIMQAKALNKPTPINLINHAYWNLGNHNSGNILDEVVQIFGSKFTPFDNN 361
Query: 204 TVPTGEIVPVKGTPFDFTSEKRIGDTINQVGL--GYDHNYVLDCG--EEKAGLRHAAKVR 259
+PTG+ VKGTP DF + +G INQ+ GY+ NYVL+ G E K GL+ AA V
Sbjct: 362 LIPTGKFSSVKGTPNDFLKPQIVGARINQLQKTNGYNVNYVLNKGKLENKEGLKVAAIVM 541
Query: 260 DPSSSRVLNLWTNAPGVQFYTGNYVDNVTGK 290
D S RV+ L TNAPG+QFY N V N GK
Sbjct: 542 DKKSGRVMKLSTNAPGLQFYRANCVKNDKGK 634
>BQ143983
Length = 738
Score = 74.3 bits (181), Expect = 6e-14
Identities = 84/233 (36%), Positives = 107/233 (45%), Gaps = 16/233 (6%)
Frame = +3
Query: 2 ADQNKKPEIFELNNGTMQLLV-TNLGCTITSFSVPAKDGVLSDVVLGLD--SVESYQKGL 58
AD NKKP IFEL NGT+ +L CTITS S AKDGVLSD V LD + S + L
Sbjct: 3 ADHNKKPNIFELTNGTIHNPW*QSLMCTITSLSAIAKDGVLSDSVHVLDIWRILSLKV*L 182
Query: 59 APYFGCIVGRVANRIKNGKFTLDGVE-YSLPLNKAPNTLHGGNVGFDKKVWDVLEYKKGE 117
+ + + +K L GV + L N P + +GFD V DVLEY
Sbjct: 183 SY*RLALTVHLQTSLKMKSAHLMGVSTHCLYTNPRPLCI-VEMLGFDNTVEDVLEYIHRR 359
Query: 118 TPSITFKY-DSHDGEEGYPGDITVTATYTLTSSTTM-RLDMEGVAKNKPTIINLAQHTYW 175
F S ATYTLT + D+ N PTIINLA TYW
Sbjct: 360 NSVYYF*V*QS*GRRRAILAT*LSPATYTLTVKARL*GSDINRSGTNMPTIINLAHFTYW 539
Query: 176 NLAGHSSGNILDHSIKISANHV------TPV---DQNTVPTGEI-VPVKGTPF 218
NL + + ++S+ IS +++ PV + P + PVKGTPF
Sbjct: 540 NL---NWP*LTEYSLLISIHNIGMN*W*LPVRS*NTRAKPW*KYRCPVKGTPF 689
>CB065399 OMNI|NTL01EC00 galactose-1-epimerase (mutarotase) {Escherichia coli
K12-MG1655}, partial (25%)
Length = 571
Score = 57.4 bits (137), Expect = 8e-09
Identities = 29/87 (33%), Positives = 45/87 (51%), Gaps = 2/87 (2%)
Frame = -2
Query: 255 AAKVRDPSSSRVLNLWTNAPGVQFYTGNYVDNVTGKGGAVYGKHAGLCLETQGFPDAVNK 314
AA V L ++T AP +QFY+GN++ +G Y GL LE++ PD+ N
Sbjct: 564 AAHVWSADEKLQLKVYTTAPALQFYSGNFLGGTPSRGTEPYADWQGLALESEFLPDSPNH 385
Query: 315 PNF--PSVVVKPGEKYQHSMLFEFSIE 339
P + P ++PGE+Y ++F E
Sbjct: 384 PEWPQPDCFLRPGEEYSSLTEYQFIAE 304
>BF520495 weakly similar to PIR|T01933|T019 probable aldose 1-epimerase (EC
5.1.3.3) - common tobacco, partial (13%)
Length = 462
Score = 46.6 bits (109), Expect = 1e-05
Identities = 21/46 (45%), Positives = 31/46 (66%)
Frame = +1
Query: 9 EIFELNNGTMQLLVTNLGCTITSFSVPAKDGVLSDVVLGLDSVESY 54
++FEL G + L VTN G T+ S +P K+G L D+VLG D+ ++Y
Sbjct: 118 KLFELKKGDLTLKVTNWGATLVSLVLPDKNGNLGDIVLGYDTPKAY 255
Score = 38.5 bits (88), Expect = 0.004
Identities = 18/41 (43%), Positives = 26/41 (62%), Gaps = 1/41 (2%)
Frame = +3
Query: 96 LHGGNVGFDKKVWDVLEY-KKGETPSITFKYDSHDGEEGYP 135
++ G GF +W V +Y ++G+ P I F Y S DGEEG+P
Sbjct: 252 IYCGPRGFSDVLWKVEKYVREGDQPLIKFGYHSFDGEEGFP 374
>TC79412 similar to GP|15809663|gb|AAL07275.1 COP9 complex subunit 6
{Arabidopsis thaliana}, partial (93%)
Length = 1380
Score = 30.4 bits (67), Expect = 1.0
Identities = 25/98 (25%), Positives = 50/98 (50%), Gaps = 2/98 (2%)
Frame = +2
Query: 111 LEYKKGETPSITFKYDSHDGEEGYPGDITVTATYTLTSSTTMRLDMEGVAKNKPTI-INL 169
+ + + + P F+ + H +G P I V ++YT+ + R+ ++ VA KP+ +
Sbjct: 554 INHSQKDLPVSIFESELHV-IDGIPQLIFVRSSYTIETVEAERISVDHVAHLKPSDGGSA 730
Query: 170 AQHTYWNLAG-HSSGNILDHSIKISANHVTPVDQNTVP 206
A NL G HS+ +L IK+ +++ +++ VP
Sbjct: 731 ATQLAANLTGIHSAIKMLHSRIKVLHHYLLAMEKGDVP 844
>TC91147
Length = 713
Score = 28.1 bits (61), Expect = 5.1
Identities = 23/88 (26%), Positives = 39/88 (44%), Gaps = 1/88 (1%)
Frame = +1
Query: 102 GFDKKVWDVLEYKKGETPSITFKYDSHD-GEEGYPGDITVTATYTLTSSTTMRLDMEGVA 160
G V D +++ + E S+T K + + G+ D T T T +TSST
Sbjct: 379 GVQSVVQDFIDFSEKEQ-SLTRKLRTDNRGKNKTSVDTTNTNTVEVTSSTNFSNGTVSEY 555
Query: 161 KNKPTIINLAQHTYWNLAGHSSGNILDH 188
K+ ++ + T+ N + G IL+H
Sbjct: 556 KSVKSVTTTSSETFVNSGATAKGAILNH 639
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.315 0.136 0.404
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,846,681
Number of Sequences: 36976
Number of extensions: 130185
Number of successful extensions: 408
Number of sequences better than 10.0: 16
Number of HSP's better than 10.0 without gapping: 397
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 397
length of query: 339
length of database: 9,014,727
effective HSP length: 97
effective length of query: 242
effective length of database: 5,428,055
effective search space: 1313589310
effective search space used: 1313589310
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 59 (27.3 bits)
Medicago: description of AC138199.6


