

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC138199.5 + phase: 0
(617 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
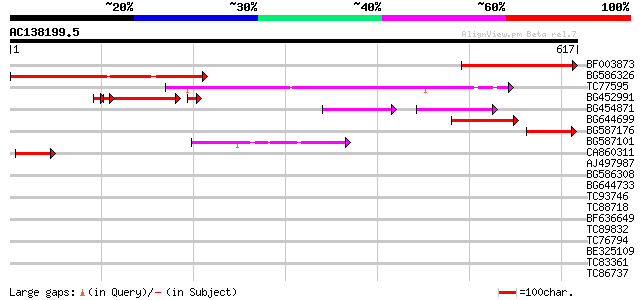
Score E
Sequences producing significant alignments: (bits) Value
BF003873 similar to GP|14715222|em putative polyprotein {Cicer a... 231 5e-61
BG586326 similar to PIR|G84493|G8 probable retroelement pol poly... 186 3e-47
TC77595 weakly similar to PIR|T18350|T18350 probable pol polypro... 179 3e-45
BG452991 PIR|A25875|A25 histone H4 - Tetrahymena thermophila, pa... 114 6e-30
BG454871 weakly similar to GP|10140673|g putative gag-pol polypr... 72 5e-17
BG644699 similar to PIR|T07863|T078 probable polyprotein - pinea... 64 1e-10
BG587176 weakly similar to PIR|G84493|G84 probable retroelement ... 56 5e-08
BG587101 similar to GP|6691191|gb F7F22.15 {Arabidopsis thaliana... 55 6e-08
CA860311 weakly similar to GP|7289872|gb|A CG17427 gene product ... 43 3e-04
AJ497987 weakly similar to GP|9927273|dbj Similar to Arabidopsis... 39 0.008
BG586308 weakly similar to PIR|F84528|F8 probable retroelement p... 37 0.017
BG644733 weakly similar to GP|15289942|db putative polyprotein {... 36 0.038
TC93746 weakly similar to GP|22830935|dbj|BAC15800. hypothetical... 31 1.2
TC88718 weakly similar to GP|3850569|gb|AAC72109.1| ESTs gb|T212... 31 1.6
BF636649 similar to GP|21628724|emb OSJNBa0033H08.7 {Oryza sativ... 30 2.1
TC89832 homologue to GP|21618319|gb|AAM67369.1 unknown {Arabidop... 30 2.7
TC76794 similar to GP|7416846|dbj|BAA94084.1 NAD-dependent sorbi... 30 3.6
BE325109 similar to PIR|T04011|T040 hypothetical protein T5L19.2... 30 3.6
TC83361 similar to PIR|E96729|E96729 unknown protein F5A18.27 [i... 29 4.7
TC86737 weakly similar to GP|6683624|dbj|BAA89272.1 Pol {Alterna... 29 4.7
>BF003873 similar to GP|14715222|em putative polyprotein {Cicer arietinum},
partial (82%)
Length = 559
Score = 231 bits (590), Expect = 5e-61
Identities = 115/126 (91%), Positives = 119/126 (94%)
Frame = +2
Query: 492 GVGRALKSKKLTPKFIGPYQILERVGTVAYRVGLPPHLSNLHNVFHVSQLQKYVPDPSHV 551
GVGRALKSKKLT +FIGPYQI ERVGTVAYRVGLPPHL NLH+VFHVSQL+KYVPDPSHV
Sbjct: 2 GVGRALKSKKLTVRFIGPYQISERVGTVAYRVGLPPHLLNLHDVFHVSQLRKYVPDPSHV 181
Query: 552 IQSDDVQVRDNLTVETLPVRIDDRKVKTLRGKEIPLVRVVWTGATGESLTWELESKMLVS 611
IQSDDVQVRDNLTVETLPVRIDDRKVKTLRGKEIPLVRVVW A GESLTWELESKM+ S
Sbjct: 182 IQSDDVQVRDNLTVETLPVRIDDRKVKTLRGKEIPLVRVVWDRANGESLTWELESKMVES 361
Query: 612 YPELFA 617
YPELFA
Sbjct: 362 YPELFA 379
>BG586326 similar to PIR|G84493|G8 probable retroelement pol polyprotein
[imported] - Arabidopsis thaliana, partial (13%)
Length = 736
Score = 186 bits (471), Expect = 3e-47
Identities = 97/215 (45%), Positives = 142/215 (65%), Gaps = 1/215 (0%)
Frame = +2
Query: 2 QEGKVVAYASRQLRVHEKNYPTHDLELAAVVFVLKIWRHYLYGSRFEVFSDHKSLKYLFD 61
Q KV+AYASRQLR HE NYPTHDLE+AAVVF LKIWR YLYG++ ++ +DHKSLKY+F
Sbjct: 98 QHEKVIAYASRQLRKHEGNYPTHDLEMAAVVFALKIWRSYLYGAKVQIHTDHKSLKYIFT 277
Query: 62 QKELNMRQRRWLELLKDYDFGLNYHPGKANVVADALSRKTLHMSAMMVREFELLEQFRDM 121
Q ELN+RQRRW+E + DYD + Y+PGKAN+VADALSR+ + +SA RE + L+
Sbjct: 278 QPELNLRQRRWMEFVADYDLDITYYPGKANLVADALSRRRVDVSA--EREADDLDGMVRA 451
Query: 122 SLVCEWSPQSVKLGMLKID-SEFLRSIKEAQKVDVKFVDLLVARDQTEDSDFKFDDQGVL 180
+ + + LG+ ++ ++ I+ AQ D + L Q + ++++ G +
Sbjct: 452 LRLNVLTKATESLGLEAVNQADLFTRIRLAQGQD----ENLQKVAQNDRTEYQTAKDGTI 619
Query: 181 RFRGRICIPDNEEIKKMILEESHRSSLSIHPGATK 215
GRI +P++ +K+ I+ E+H+S S+HPGA +
Sbjct: 620 LVNGRISVPNDRSLKEEIMSEAHKSRFSVHPGAPR 724
>TC77595 weakly similar to PIR|T18350|T18350 probable pol polyprotein - rice
blast fungus gypsy retroelement (fragment), partial
(14%)
Length = 1708
Score = 179 bits (454), Expect = 3e-45
Identities = 124/392 (31%), Positives = 193/392 (48%), Gaps = 13/392 (3%)
Frame = +2
Query: 170 SDFKFDDQGVLRFRGRICIPDNE-------EIKKMILEESHRSSLSIHPGATKMYHDLKK 222
S+ + D L FRGRI +P ++ E++ +++ESH S+ + HPG + +
Sbjct: 74 SECQLDSLKRLTFRGRIWVPGSDDEESPLNELRTKLVQESHDSTAAGHPGRNGTLEIVSR 253
Query: 223 IFWWSGLKRDVAQFVYSCLVCQKSKVEHQKPAGMMVPLDVPEWKWDSISMDFVTSLPNTP 282
F+W G + V +FV +C VC + Q G + PL VP +SMDF+TSLP T
Sbjct: 254 KFFWPGQSQTVRRFVRNCDVCGGIHIWRQAKRGFLKPLPVPNRLHSDLSMDFITSLPPTR 433
Query: 283 -RGNDAIWVIVDRLTKSAHFLPINISFPVAQLAEIYIKEIVKLHGVPSSIVSDRDPRFTS 341
RG+ +WVIVDRL+KS ++ + A+ ++ + HG+P SIVSDR +
Sbjct: 434 GRGSQYLWVIVDRLSKSVTLEEMD-TMEAEACAQRFLSCHYRFHGMPQSIVSDRGSNWVG 610
Query: 342 RFWKSLQEALGSKLRLSSAYHPQTDGQSERTIQSLEDLLRICVLEQGGTWDSHLPLIEFT 401
RFW+ G LS++YHPQTDG +ER Q ++ +LR V W LP ++
Sbjct: 611 RFWREFCRLTGVTQLLSTSYHPQTDGGTERWNQEIQAVLRAYVCWSQDNWGDLLPTVQLA 790
Query: 402 YNNSYHSSIGMAPFEALYGRRCRIPLCWFESGERVVLGPAIVQQTTEKVQ----MIQEKI 457
N ++SSIG PF +G ++G V G A Q ++++ IQ +I
Sbjct: 791 LRNRHNSSIGATPFFVEHGYHVDPIPTVEDTGGVVSEGEAAAQLLVKRMKDVTGFIQAEI 970
Query: 458 KASQSRQKTYHDKRRKDLE-FQEGDHVFLRVTPMTGVGRALKSKKLTPKFIGPYQILERV 516
A+Q R + +KRR + +Q GD V+L V+ + K L K Y++ V
Sbjct: 971 VAAQQRSEASANKRRCPADRYQVGDKVWLNVSNYKSPRPSKKLDWLHHK----YEVTRFV 1138
Query: 517 GTVAYRVGLPPHLSNLHNVFHVSQLQKYVPDP 548
+ +P ++ FHV L++ DP
Sbjct: 1139TPHVVELNVP---GTVYPKFHVDLLRRAASDP 1225
>BG452991 PIR|A25875|A25 histone H4 - Tetrahymena thermophila, partial (33%)
Length = 560
Score = 114 bits (284), Expect(4) = 6e-30
Identities = 57/78 (73%), Positives = 65/78 (83%)
Frame = +3
Query: 109 VREFELLEQFRDMSLVCEWSPQSVKLGMLKIDSEFLRSIKEAQKVDVKFVDLLVARDQTE 168
+ + LEQFRD+SLVCE SPQSVKLGMLKI++EFL SIKEAQKVDVK VDL+ +QTE
Sbjct: 51 LESWSCLEQFRDLSLVCEVSPQSVKLGMLKINNEFLDSIKEAQKVDVKLVDLMFGNNQTE 230
Query: 169 DSDFKFDDQGVLRFRGRI 186
D DFK DDQGVL+FR RI
Sbjct: 231 DGDFKVDDQGVLQFRDRI 284
Score = 26.9 bits (58), Expect(4) = 6e-30
Identities = 11/15 (73%), Positives = 15/15 (99%)
Frame = +2
Query: 194 IKKMILEESHRSSLS 208
+KKMILEESHRS+++
Sbjct: 284 MKKMILEESHRSNVN 328
Score = 25.4 bits (54), Expect(4) = 6e-30
Identities = 11/13 (84%), Positives = 12/13 (91%)
Frame = +1
Query: 92 VVADALSRKTLHM 104
VVAD LSRKTLH+
Sbjct: 1 VVADVLSRKTLHV 39
Score = 23.1 bits (48), Expect(4) = 6e-30
Identities = 10/12 (83%), Positives = 11/12 (91%)
Frame = +2
Query: 103 HMSAMMVREFEL 114
+MSAMMVRE EL
Sbjct: 32 YMSAMMVRELEL 67
>BG454871 weakly similar to GP|10140673|g putative gag-pol polyprotein {Oryza
sativa (japonica cultivar-group)}, partial (7%)
Length = 674
Score = 72.4 bits (176), Expect(2) = 5e-17
Identities = 37/81 (45%), Positives = 46/81 (56%)
Frame = +2
Query: 341 SRFWKSLQEALGSKLRLSSAYHPQTDGQSERTIQSLEDLLRICVLEQGGTWDSHLPLIEF 400
S FWK L + G+ L +SSAYHP +DGQSE + E LR + W P E+
Sbjct: 32 SNFWKQLFKLHGTILTMSSAYHP*SDGQSEALNKGXEMYLRCLMFTDPLKWSKAFPWAEY 211
Query: 401 TYNNSYHSSIGMAPFEALYGR 421
YN SY+ S M PF+ALYGR
Sbjct: 212 WYNTSYNISAAMTPFKALYGR 274
Score = 33.5 bits (75), Expect(2) = 5e-17
Identities = 26/92 (28%), Positives = 43/92 (46%), Gaps = 4/92 (4%)
Frame = +1
Query: 443 VQQTTEKVQMIQEKIKASQSRQKTYH--DKRRKDLEFQEGDHVFLRVTPMTGVGRALK-- 498
V +TE+ + I +++Q H DK+R+ EFQ G+HV +++ P AL+
Sbjct: 331 VPVSTERRIAVTATIHLYKAQQTMKHQADKKRRHFEFQLGEHVLVKLQPYQQSSVALRKY 510
Query: 499 SKKLTPKFIGPYQILERVGTVAYRVGLPPHLS 530
K +P F + A+ PP+LS
Sbjct: 511 QKFGSPNFGSLLTVCSL*VESAFHCKSPPYLS 606
>BG644699 similar to PIR|T07863|T078 probable polyprotein - pineapple
retrotransposon dea1 (fragment), partial (5%)
Length = 231
Score = 64.3 bits (155), Expect = 1e-10
Identities = 31/74 (41%), Positives = 50/74 (66%), Gaps = 1/74 (1%)
Frame = +2
Query: 481 DHVFLRVTPMT-GVGRALKSKKLTPKFIGPYQILERVGTVAYRVGLPPHLSNLHNVFHVS 539
+ V L+V P G R K KL+ ++IGP+++++R+G VAY + LPP LS +H VFHVS
Sbjct: 2 EQVLLKVLPTERGDCRFGKRGKLSLRYIGPFEVIKRIGEVAYELALPPGLSGVHPVFHVS 181
Query: 540 QLQKYVPDPSHVIQ 553
++Y D +++I+
Sbjct: 182 MFKRYHGDGNYIIR 223
>BG587176 weakly similar to PIR|G84493|G84 probable retroelement pol
polyprotein [imported] - Arabidopsis thaliana, partial
(1%)
Length = 729
Score = 55.8 bits (133), Expect = 5e-08
Identities = 26/54 (48%), Positives = 35/54 (64%)
Frame = -1
Query: 563 LTVETLPVRIDDRKVKTLRGKEIPLVRVVWTGATGESLTWELESKMLVSYPELF 616
L +ET PVRI DR K +R K I +V++VW + E +TWE E++M YPE F
Sbjct: 717 LDLETRPVRILDRMEKAMRKKPIQMVKIVWDCSGREEITWETEARMKADYPEWF 556
>BG587101 similar to GP|6691191|gb F7F22.15 {Arabidopsis thaliana}, partial
(10%)
Length = 624
Score = 55.5 bits (132), Expect = 6e-08
Identities = 44/179 (24%), Positives = 86/179 (47%), Gaps = 6/179 (3%)
Frame = +2
Query: 198 ILEESHRSSLSIHPGATKMYHDLKKI-FWWSGLKRDVAQFVYSCLVCQKS---KVEHQKP 253
IL H S+ + H +K +++ FWW + +D F+ C CQ+ ++ P
Sbjct: 104 ILFHCHGSNYAGHFAVSKTVSKIQQAGFWWPTMFKDAHSFISKCDPCQRQGNIS*RNEMP 283
Query: 254 AGMMVPLDVPEWKWDSISMDFVTSLPNTPRGNDAIWVIVDRLTKSAHFL--PINISFPVA 311
++ ++V +D +DF+ P++ N I V VD ++K + P N + V
Sbjct: 284 QNFILEVEV----FDVWGIDFMGPFPSS-YNNKYILVAVDYVSKWVEAIASPTNDATVVV 448
Query: 312 QLAEIYIKEIVKLHGVPSSIVSDRDPRFTSRFWKSLQEALGSKLRLSSAYHPQTDGQSE 370
++ + I GVP ++SD F ++ ++ L + G + ++++AYHPQ +S+
Sbjct: 449 KM---FKSVIFPRFGVPRVVISDGGSHFINKVFEKLLKKNGVRHKVATAYHPQKAERSK 616
>CA860311 weakly similar to GP|7289872|gb|A CG17427 gene product {Drosophila
melanogaster}, partial (20%)
Length = 192
Score = 43.1 bits (100), Expect = 3e-04
Identities = 18/43 (41%), Positives = 29/43 (66%)
Frame = +1
Query: 7 VAYASRQLRVHEKNYPTHDLELAAVVFVLKIWRHYLYGSRFEV 49
+AYASR L E+NY + E A ++ ++ +RHYL+G +FE+
Sbjct: 64 IAYASRLLTAAERNYTVVERECLAAIWAIRNFRHYLHGPKFEL 192
>AJ497987 weakly similar to GP|9927273|dbj Similar to Arabidopsis thaliana
chromosome II BAC F26H6; putative retroelement pol
polyprotein, partial (1%)
Length = 636
Score = 38.5 bits (88), Expect = 0.008
Identities = 24/76 (31%), Positives = 39/76 (50%), Gaps = 3/76 (3%)
Frame = -2
Query: 30 AVVFVLKIWRHYLYGSRFEVFSDHKSLKYLFDQKELNMRQRRWLELLKDYDFGLNYHPGK 89
A+ + K RHY+ + S +KY+F++ L R RW LL +YD + Y K
Sbjct: 623 ALAWAAKRLRHYMINHTTWLVSKMDPIKYIFEKPALTGRIARWQMLLSEYD--IEYRSQK 450
Query: 90 A---NVVADALSRKTL 102
A +++AD L+ + L
Sbjct: 449 AIKGSILADHLAHQPL 402
>BG586308 weakly similar to PIR|F84528|F8 probable retroelement pol
polyprotein [imported] - Arabidopsis thaliana, partial
(7%)
Length = 686
Score = 37.4 bits (85), Expect = 0.017
Identities = 21/71 (29%), Positives = 37/71 (51%)
Frame = -2
Query: 325 HGVPSSIVSDRDPRFTSRFWKSLQEALGSKLRLSSAYHPQTDGQSERTIQSLEDLLRICV 384
HG+P IV+D F S ++ E +L +S +PQ++GQ+E + + + D L+ +
Sbjct: 685 HGLPYEIVTDNGSHFISNKFREFCERWRIRLNTASPRYPQSNGQAEASNKIIIDGLKKRL 506
Query: 385 LEQGGTWDSHL 395
+ G W L
Sbjct: 505 DLKKGCWADEL 473
>BG644733 weakly similar to GP|15289942|db putative polyprotein {Oryza sativa
(japonica cultivar-group)}, partial (1%)
Length = 174
Score = 36.2 bits (82), Expect = 0.038
Identities = 18/40 (45%), Positives = 25/40 (62%)
Frame = -3
Query: 286 DAIWVIVDRLTKSAHFLPINISFPVAQLAEIYIKEIVKLH 325
++I V+VDRLTKS F+P S+ A I + EIV +H
Sbjct: 121 ESI*VVVDRLTKSTLFIPFKTSYSAK*YARILLDEIVCIH 2
>TC93746 weakly similar to GP|22830935|dbj|BAC15800. hypothetical
protein~similar to gag-pol polyprotein {Oryza sativa
(japonica cultivar-group)}, partial (4%)
Length = 1019
Score = 31.2 bits (69), Expect = 1.2
Identities = 13/41 (31%), Positives = 25/41 (60%)
Frame = +2
Query: 262 VPEWKWDSISMDFVTSLPNTPRGNDAIWVIVDRLTKSAHFL 302
VP+ W+ +++DF L T + D+ V+ D+ ++ AHF+
Sbjct: 476 VPKPPWEDVTIDFSLGLL*TQQLKDSKMVVGDKFSRMAHFI 598
>TC88718 weakly similar to GP|3850569|gb|AAC72109.1| ESTs gb|T21276
gb|T45403 and gb|AA586113 come from this gene.
{Arabidopsis thaliana}, partial (16%)
Length = 1356
Score = 30.8 bits (68), Expect = 1.6
Identities = 18/53 (33%), Positives = 26/53 (48%), Gaps = 8/53 (15%)
Frame = +3
Query: 132 VKLGMLKIDSEF--------LRSIKEAQKVDVKFVDLLVARDQTEDSDFKFDD 176
VK+ KID E L +K+A+ KFV ++ D+ ED + FDD
Sbjct: 747 VKIVPSKIDDEIFWSRYFYKLHKLKQAEDARAKFVKRAISGDEEEDLSWDFDD 905
>BF636649 similar to GP|21628724|emb OSJNBa0033H08.7 {Oryza sativa}, partial
(4%)
Length = 653
Score = 30.4 bits (67), Expect = 2.1
Identities = 17/55 (30%), Positives = 26/55 (46%)
Frame = +1
Query: 365 TDGQSERTIQSLEDLLRICVLEQGGTWDSHLPLIEFTYNNSYHSSIGMAPFEALY 419
+D Q+ +LE L EQ G + L E YN ++H + G PF+ +Y
Sbjct: 7 SDEQAGLLNHTLETHLLYFTSEQQGV*NFFLTWAECLYNTNFHRTAGCTPFKVVY 171
>TC89832 homologue to GP|21618319|gb|AAM67369.1 unknown {Arabidopsis
thaliana}, partial (62%)
Length = 1347
Score = 30.0 bits (66), Expect = 2.7
Identities = 18/49 (36%), Positives = 23/49 (46%)
Frame = +1
Query: 323 KLHGVPSSIVSDRDPRFTSRFWKSLQEALGSKLRLSSAYHPQTDGQSER 371
+++G PS I R FW S+ LGS R Y P TDG + R
Sbjct: 727 QIYGPPSKINKARQRTNVQVFWGSVH-GLGSFCRRCYCYSPDTDGIARR 870
>TC76794 similar to GP|7416846|dbj|BAA94084.1 NAD-dependent sorbitol
dehydrogenase {Prunus persica}, partial (93%)
Length = 1691
Score = 29.6 bits (65), Expect = 3.6
Identities = 12/21 (57%), Positives = 17/21 (80%)
Frame = -2
Query: 530 SNLHNVFHVSQLQKYVPDPSH 550
SNL N+FHVS + YVP+P++
Sbjct: 1690 SNLINIFHVS*FKVYVPNPTN 1628
>BE325109 similar to PIR|T04011|T040 hypothetical protein T5L19.200 -
Arabidopsis thaliana, partial (9%)
Length = 430
Score = 29.6 bits (65), Expect = 3.6
Identities = 14/43 (32%), Positives = 25/43 (57%)
Frame = +1
Query: 345 KSLQEALGSKLRLSSAYHPQTDGQSERTIQSLEDLLRICVLEQ 387
KSLQ G++++L + P+ D ERT+Q D +I + ++
Sbjct: 79 KSLQTKTGARIQLIPQHLPEGDDSKERTVQVTGDKRQIEIAQE 207
>TC83361 similar to PIR|E96729|E96729 unknown protein F5A18.27 [imported] -
Arabidopsis thaliana, partial (57%)
Length = 1095
Score = 29.3 bits (64), Expect = 4.7
Identities = 44/165 (26%), Positives = 67/165 (39%), Gaps = 7/165 (4%)
Frame = +2
Query: 330 SIVSDR-DPRFTSRFWKSLQEALGSKLRLSSAYHPQTDGQSERTIQSLEDLLRICVLEQG 388
SIVSD P T++ +K+ E L KLR + + + + +TIQS + L C+L
Sbjct: 284 SIVSDSIHPSKTNQTYKAGSELL--KLRRIRTHLMKINKPAVKTIQSPDGDLIDCILSHH 457
Query: 389 GT------WDSHLPLIEFTYNNSYHSSIGMAPFEALYGRRCRIPLCWFESGERVVLGPAI 442
H PL N Y+++ G E L W +SGE G
Sbjct: 458 QPAFDHPKLKGHKPLDPPERPNGYYNN-GEKVSERLQ--------LWTDSGEECPEGTVP 610
Query: 443 VQQTTEKVQMIQEKIKASQSRQKTYHDKRRKDLEFQEGDHVFLRV 487
+++TTE + IK + K R+D + +H L V
Sbjct: 611 IRRTTEDDILRASSIKRFGRKPKPV----RRDSTSSDHEHAILFV 733
>TC86737 weakly similar to GP|6683624|dbj|BAA89272.1 Pol {Alternaria
alternata}, partial (21%)
Length = 1540
Score = 29.3 bits (64), Expect = 4.7
Identities = 14/25 (56%), Positives = 17/25 (68%)
Frame = +1
Query: 7 VAYASRQLRVHEKNYPTHDLELAAV 31
VA+ S++L E NYP HD EL AV
Sbjct: 1438 VAFHSQRLSPAEYNYPIHDKELLAV 1512
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.321 0.137 0.411
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 19,301,355
Number of Sequences: 36976
Number of extensions: 270427
Number of successful extensions: 1343
Number of sequences better than 10.0: 42
Number of HSP's better than 10.0 without gapping: 1336
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1339
length of query: 617
length of database: 9,014,727
effective HSP length: 102
effective length of query: 515
effective length of database: 5,243,175
effective search space: 2700235125
effective search space used: 2700235125
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 61 (28.1 bits)
Medicago: description of AC138199.5


