

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC138199.3 - phase: 0
(306 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
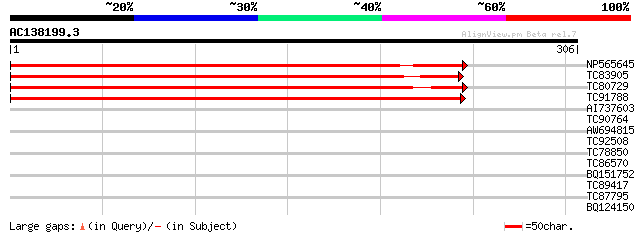
Score E
Sequences producing significant alignments: (bits) Value
NP565645 NP565645|AY115840.1|AAM55301.1 auxin efflux carrier pro... 410 e-115
TC83905 auxin efflux carrier protein [Medicago truncatula] 371 e-103
TC80729 auxin efflux carrier protein [Medicago truncatula] 369 e-103
TC91788 auxin efflux carrier protein [Medicago truncatula] 345 1e-95
AI737603 30 1.2
TC90764 similar to PIR|T05666|T05666 hypothetical protein F22I13... 28 3.4
AW694815 similar to GP|22091415|gb putative RNA helicase {Arabid... 28 5.8
TC92508 weakly similar to GP|5091614|gb|AAD39602.1| Contains a P... 27 7.5
TC78850 homologue to GP|10177413|dbj|BAB10544. fibrillarin-like ... 27 7.5
TC86570 homologue to GP|11094365|gb|AAG29593.1 Ser/Thr specific ... 27 7.5
BQ151752 homologue to PIR|T09555|T09 fibrillarin - Arabidopsis t... 27 7.5
TC89417 similar to PIR|F96736|F96736 unknown protein F23N20.18 [... 27 9.8
TC87795 similar to GP|14164407|dbj|BAB55806. cytosolic aldehyde ... 27 9.8
BQ124150 weakly similar to PIR|T45884|T458 hypothetical protein ... 27 9.8
>NP565645 NP565645|AY115840.1|AAM55301.1 auxin efflux carrier protein
[Medicago truncatula]
Length = 1575
Score = 410 bits (1055), Expect = e-115
Identities = 212/249 (85%), Positives = 223/249 (89%), Gaps = 2/249 (0%)
Frame = +1
Query: 1 MITLTDFYHVMTAMVPLYVAMILAYGSVKWWKIFSPDQCSGINRFVALFAVPLLSFHFIA 60
MITLTDFYHVMT+MVPLYVAMILAYGSVKWWKIFSPDQCSGINRFVALFAVPLLSFHFIA
Sbjct: 1 MITLTDFYHVMTSMVPLYVAMILAYGSVKWWKIFSPDQCSGINRFVALFAVPLLSFHFIA 180
Query: 61 SNNPYKMNLRFLAADTLQKLIILCLLAIWSNFSKRGCLEWTITLFSLSTLPNTLVMGIPL 120
SNNPYKMN RFL ADTLQK+I+L L IW+N SKRGCLEWTITLFS+STLPNTLVMGIPL
Sbjct: 181 SNNPYKMNTRFLVADTLQKIIVLLALTIWANVSKRGCLEWTITLFSISTLPNTLVMGIPL 360
Query: 121 LKGMY-GEFSGSLMVQIVVLQCIIWYTLMLFMFEFRGARLLIFEQFPDTAGSIVSIHVDS 179
LK MY GEFSGSLMVQIVVLQCIIWYTL+LFMFEFRGAR+LI EQFPDTAG+IVSIHVDS
Sbjct: 361 LKCMYGGEFSGSLMVQIVVLQCIIWYTLILFMFEFRGARMLISEQFPDTAGTIVSIHVDS 540
Query: 180 DVMSL-DGRTPLETDAEIKEDGKLHITVRKSNASRSDIYSRRSQGLSSNTPRPSNLTNAE 238
DVMSL DGR LET+A IKEDGKLH+ VR+SNA SRRS S TPR SNLTNAE
Sbjct: 541 DVMSLDDGRQCLETEAVIKEDGKLHVIVRRSNA------SRRSHSQSHTTPRASNLTNAE 702
Query: 239 IYSLQSSRN 247
IYSLQSSRN
Sbjct: 703 IYSLQSSRN 729
>TC83905 auxin efflux carrier protein [Medicago truncatula]
Length = 1869
Score = 371 bits (952), Expect = e-103
Identities = 183/245 (74%), Positives = 211/245 (85%)
Frame = +1
Query: 1 MITLTDFYHVMTAMVPLYVAMILAYGSVKWWKIFSPDQCSGINRFVALFAVPLLSFHFIA 60
MITL D Y V+TA+VPLYVAMILAYGSV+WWKIFSPDQCSGINRFVA+FAVPLLSFHFI+
Sbjct: 1 MITLKDLYTVLTAVVPLYVAMILAYGSVRWWKIFSPDQCSGINRFVAVFAVPLLSFHFIS 180
Query: 61 SNNPYKMNLRFLAADTLQKLIILCLLAIWSNFSKRGCLEWTITLFSLSTLPNTLVMGIPL 120
+NNPY+MN RF+AADTLQK+I+L L++W+ F+K G LEW IT+FSLSTLPNTLVMGIPL
Sbjct: 181 TNNPYQMNFRFIAADTLQKIIMLVALSLWTLFTKNGNLEWMITIFSLSTLPNTLVMGIPL 360
Query: 121 LKGMYGEFSGSLMVQIVVLQCIIWYTLMLFMFEFRGARLLIFEQFPDTAGSIVSIHVDSD 180
L MYG++SGSLMVQ+VVLQCIIWYTL+LF+FE+RGA+LLI EQFP+TA SIVS VDSD
Sbjct: 361 LIAMYGDYSGSLMVQVVVLQCIIWYTLLLFLFEYRGAKLLIMEQFPETAASIVSFKVDSD 540
Query: 181 VMSLDGRTPLETDAEIKEDGKLHITVRKSNASRSDIYSRRSQGLSSNTPRPSNLTNAEIY 240
VMSLDGR LETDA + +DGKLH+TVRKSNASR + NTPRPSNLT AEIY
Sbjct: 541 VMSLDGRDFLETDASVGDDGKLHVTVRKSNASR--------RSFMMNTPRPSNLTGAEIY 696
Query: 241 SLQSS 245
SL S+
Sbjct: 697 SLSST 711
>TC80729 auxin efflux carrier protein [Medicago truncatula]
Length = 2366
Score = 369 bits (947), Expect = e-103
Identities = 183/247 (74%), Positives = 208/247 (84%)
Frame = +1
Query: 1 MITLTDFYHVMTAMVPLYVAMILAYGSVKWWKIFSPDQCSGINRFVALFAVPLLSFHFIA 60
MIT D Y V+TA+VPLYVAMILAYGSV+WWKIFSPDQCSGINRFVA+FAVPLLSFHFI+
Sbjct: 1 MITWHDLYTVLTAVVPLYVAMILAYGSVRWWKIFSPDQCSGINRFVAIFAVPLLSFHFIS 180
Query: 61 SNNPYKMNLRFLAADTLQKLIILCLLAIWSNFSKRGCLEWTITLFSLSTLPNTLVMGIPL 120
+NNPY MN RF+AADTLQK+I+L L IW+NF+ G LEW IT+FSLSTLPNTLVMGIPL
Sbjct: 181 TNNPYAMNYRFIAADTLQKIIMLLALTIWTNFTANGSLEWMITIFSLSTLPNTLVMGIPL 360
Query: 121 LKGMYGEFSGSLMVQIVVLQCIIWYTLMLFMFEFRGARLLIFEQFPDTAGSIVSIHVDSD 180
L MYGE+SG+LMVQ+VVLQCIIWYTL+LF+FE+RGA+LLI EQFP+TA SI+S VDSD
Sbjct: 361 LIAMYGEYSGTLMVQVVVLQCIIWYTLLLFLFEYRGAKLLIMEQFPETAASIISFKVDSD 540
Query: 181 VMSLDGRTPLETDAEIKEDGKLHITVRKSNASRSDIYSRRSQGLSSNTPRPSNLTNAEIY 240
V+SLDGR LETDAE+ +DGKLH+ VRKSNASR TPRPSNLT AEIY
Sbjct: 541 VVSLDGRDFLETDAEVGDDGKLHVHVRKSNASRRSFMM---------TPRPSNLTGAEIY 693
Query: 241 SLQSSRN 247
SL SSRN
Sbjct: 694 SLSSSRN 714
>TC91788 auxin efflux carrier protein [Medicago truncatula]
Length = 2076
Score = 345 bits (886), Expect = 1e-95
Identities = 174/247 (70%), Positives = 204/247 (82%), Gaps = 1/247 (0%)
Frame = +1
Query: 1 MITLTDFYHVMTAMVPLYVAMILAYGSVKWWKIFSPDQCSGINRFVALFAVPLLSFHFIA 60
MIT D Y V A+VPLYVAMILAYGSV+WWKIF+PDQCSGINRFVA+FAVPLLSFHFI+
Sbjct: 1 MITGKDIYDVFAAIVPLYVAMILAYGSVRWWKIFTPDQCSGINRFVAVFAVPLLSFHFIS 180
Query: 61 SNNPYKMNLRFLAADTLQKLIILCLLAIWSNFSK-RGCLEWTITLFSLSTLPNTLVMGIP 119
SN+PY MN FLAAD+LQK++IL L IW+ FSK + L+WTITLFSLSTLPNTLVMGIP
Sbjct: 181 SNDPYAMNYHFLAADSLQKVVILGALFIWNTFSKNQDSLDWTITLFSLSTLPNTLVMGIP 360
Query: 120 LLKGMYGEFSGSLMVQIVVLQCIIWYTLMLFMFEFRGARLLIFEQFPDTAGSIVSIHVDS 179
LL+ MYG+FSG+LMVQIVVLQ +IWYTLMLF+FE+R A+LLI EQFP+TA SI S VDS
Sbjct: 361 LLRAMYGDFSGNLMVQIVVLQSVIWYTLMLFLFEYRAAKLLITEQFPETAASITSFKVDS 540
Query: 180 DVMSLDGRTPLETDAEIKEDGKLHITVRKSNASRSDIYSRRSQGLSSNTPRPSNLTNAEI 239
DV+SL+GR PL+TDAEI EDGKLH+ V++S + S L++ TPR SNLT EI
Sbjct: 541 DVVSLNGREPLQTDAEIGEDGKLHVVVKRSTTNSMVSGSFNKSHLNNMTPRASNLTGVEI 720
Query: 240 YSLQSSR 246
YS+QSSR
Sbjct: 721 YSVQSSR 741
>AI737603
Length = 531
Score = 30.0 bits (66), Expect = 1.2
Identities = 14/22 (63%), Positives = 14/22 (63%)
Frame = +1
Query: 284 TLKHTLSTHSNNMLTHHPHPLF 305
TL HT THSN L HHP LF
Sbjct: 139 TL*HTHLTHSNKKLPHHPFFLF 204
>TC90764 similar to PIR|T05666|T05666 hypothetical protein F22I13.150 -
Arabidopsis thaliana, partial (57%)
Length = 1219
Score = 28.5 bits (62), Expect = 3.4
Identities = 21/64 (32%), Positives = 29/64 (44%)
Frame = +2
Query: 35 SPDQCSGINRFVALFAVPLLSFHFIASNNPYKMNLRFLAADTLQKLIILCLLAIWSNFSK 94
+P C GI A+F PLL ++F K+ + A T+ I LL IW +K
Sbjct: 773 TPVICLGIGNLSAVFLFPLLMYYF-------KLGVAGAAISTVLSQYIGTLLMIWC-LNK 928
Query: 95 RGCL 98
R L
Sbjct: 929 RAVL 940
>AW694815 similar to GP|22091415|gb putative RNA helicase {Arabidopsis
thaliana}, partial (16%)
Length = 404
Score = 27.7 bits (60), Expect = 5.8
Identities = 9/18 (50%), Positives = 14/18 (77%)
Frame = -1
Query: 31 WKIFSPDQCSGINRFVAL 48
WKI +P QC+G N+++ L
Sbjct: 233 WKIHTPSQCAGTNQYLNL 180
>TC92508 weakly similar to GP|5091614|gb|AAD39602.1| Contains a PF|00561
alpha/beta hydrolase fold domain. {Arabidopsis
thaliana}, partial (27%)
Length = 731
Score = 27.3 bits (59), Expect = 7.5
Identities = 28/106 (26%), Positives = 43/106 (40%), Gaps = 7/106 (6%)
Frame = +2
Query: 92 FSKRGCLEWTITLFSLSTLPNTLVMGIPLLKGMYG--EFSGSLMVQIVVLQCI-----IW 144
FS C + S S + + +V P+L+G Y + S +Q + L
Sbjct: 188 FSGFSCSPAEFSYNSCSQIYDAVVSKCPILQGGYSVTPWLSSPHLQTIFLNFFGNAPSFK 367
Query: 145 YTLMLFMFEFRGARLLIFEQFPDTAGSIVSIHVDSDVMSLDGRTPL 190
Y LF G L + D +GS+ H D DV++ D TP+
Sbjct: 368 YKRQLFTTSDGGTLALDWVTSSDVSGSV---HHDDDVVTKDDSTPI 496
>TC78850 homologue to GP|10177413|dbj|BAB10544. fibrillarin-like
{Arabidopsis thaliana}, partial (87%)
Length = 1177
Score = 27.3 bits (59), Expect = 7.5
Identities = 12/38 (31%), Positives = 24/38 (62%)
Frame = -3
Query: 48 LFAVPLLSFHFIASNNPYKMNLRFLAADTLQKLIILCL 85
LF++ L H +++NN + L++ +D L+ L ++CL
Sbjct: 977 LFSIFFLLRHSVSTNNTSMVPLKWFESDLLKWLKLICL 864
>TC86570 homologue to GP|11094365|gb|AAG29593.1 Ser/Thr specific protein
phosphatase 2A A regulatory subunit alpha isoform,
partial (35%)
Length = 883
Score = 27.3 bits (59), Expect = 7.5
Identities = 13/28 (46%), Positives = 18/28 (63%)
Frame = -2
Query: 219 RRSQGLSSNTPRPSNLTNAEIYSLQSSR 246
+R+ LSS P P+ L N IYS+ +SR
Sbjct: 192 QRAPNLSSKNPTPN*LANKGIYSIIASR 109
>BQ151752 homologue to PIR|T09555|T09 fibrillarin - Arabidopsis thaliana,
partial (53%)
Length = 682
Score = 27.3 bits (59), Expect = 7.5
Identities = 12/38 (31%), Positives = 24/38 (62%)
Frame = -3
Query: 48 LFAVPLLSFHFIASNNPYKMNLRFLAADTLQKLIILCL 85
LF++ L H +++NN + L++ +D L+ L ++CL
Sbjct: 542 LFSIFFLLRHSVSTNNTSMVPLKWFESDLLKWLKLICL 429
>TC89417 similar to PIR|F96736|F96736 unknown protein F23N20.18 [imported] -
Arabidopsis thaliana, partial (77%)
Length = 1077
Score = 26.9 bits (58), Expect = 9.8
Identities = 12/45 (26%), Positives = 27/45 (59%)
Frame = +1
Query: 11 MTAMVPLYVAMILAYGSVKWWKIFSPDQCSGINRFVALFAVPLLS 55
M +++PL +A ++ S+ +W+ F + + +FV A+PL++
Sbjct: 325 MISIIPLVLAGVI---SIVYWRFFDDLRPYALIQFVPCIAIPLMA 450
>TC87795 similar to GP|14164407|dbj|BAB55806. cytosolic aldehyde
dehydrogenase {Oryza sativa (japonica cultivar-group)},
partial (92%)
Length = 1788
Score = 26.9 bits (58), Expect = 9.8
Identities = 15/34 (44%), Positives = 20/34 (58%)
Frame = +3
Query: 165 FPDTAGSIVSIHVDSDVMSLDGRTPLETDAEIKE 198
F TAG+ VS H+D D +S G T +T EI +
Sbjct: 780 FGATAGAAVSSHMDIDAVSFTGST--QTGREIMQ 875
>BQ124150 weakly similar to PIR|T45884|T458 hypothetical protein F4P12.140 -
Arabidopsis thaliana, partial (7%)
Length = 665
Score = 26.9 bits (58), Expect = 9.8
Identities = 11/21 (52%), Positives = 18/21 (85%)
Frame = +3
Query: 219 RRSQGLSSNTPRPSNLTNAEI 239
RR+ GLSS+TP S+L+++E+
Sbjct: 261 RRAYGLSSSTPMSSSLSSSEL 323
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.325 0.137 0.415
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,457,292
Number of Sequences: 36976
Number of extensions: 126935
Number of successful extensions: 1174
Number of sequences better than 10.0: 28
Number of HSP's better than 10.0 without gapping: 1154
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1168
length of query: 306
length of database: 9,014,727
effective HSP length: 96
effective length of query: 210
effective length of database: 5,465,031
effective search space: 1147656510
effective search space used: 1147656510
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 58 (26.9 bits)
Medicago: description of AC138199.3


