

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC138133.8 + phase: 0 /pseudo
(751 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
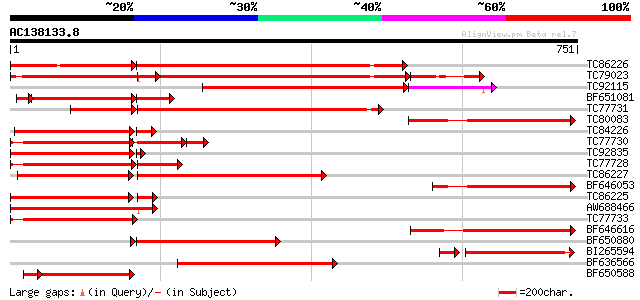
Score E
Sequences producing significant alignments: (bits) Value
TC86226 weakly similar to GP|18033111|gb|AAL56987.1 functional c... 434 0.0
TC79023 weakly similar to GP|18033111|gb|AAL56987.1 functional c... 369 e-122
TC92115 weakly similar to GP|18033111|gb|AAL56987.1 functional c... 292 e-103
BF651081 weakly similar to GP|18033111|g functional candidate re... 251 4e-99
TC77731 weakly similar to GP|18033111|gb|AAL56987.1 functional c... 339 2e-93
TC80083 weakly similar to GP|18033111|gb|AAL56987.1 functional c... 305 4e-83
TC84226 similar to GP|22037313|gb|AAM89998.1 disease resistance-... 264 3e-79
TC77730 weakly similar to GP|7341113|gb|AAF61210.1| unknown {Gly... 232 5e-78
TC92835 weakly similar to GP|18033111|gb|AAL56987.1 functional c... 278 3e-76
TC77728 similar to GP|12056928|gb|AAG48132.1 putative resistance... 235 8e-76
TC86227 weakly similar to GP|18033111|gb|AAL56987.1 functional c... 276 3e-74
BF646053 weakly similar to GP|18033111|g functional candidate re... 275 6e-74
TC86225 weakly similar to GP|9858476|gb|AAG01051.1| resistance p... 247 5e-69
AW688466 similar to GP|9965109|gb| resistance protein MG13 {Glyc... 250 2e-66
TC77733 weakly similar to GP|18033111|gb|AAL56987.1 functional c... 244 9e-65
BF646616 similar to GP|9965103|gb|A resistance protein LM6 {Glyc... 238 5e-63
BF650880 weakly similar to GP|9858476|gb| resistance protein LM1... 235 4e-62
BI265594 weakly similar to GP|9965103|gb| resistance protein LM6... 215 1e-60
BF636566 weakly similar to GP|9965103|gb| resistance protein LM6... 229 2e-60
BF650588 weakly similar to GP|7341113|gb|A unknown {Glycine max}... 202 9e-59
>TC86226 weakly similar to GP|18033111|gb|AAL56987.1 functional candidate
resistance protein KR1 {Glycine max}, partial (24%)
Length = 1769
Score = 434 bits (1115), Expect(2) = 0.0
Identities = 243/361 (67%), Positives = 278/361 (76%), Gaps = 1/361 (0%)
Frame = +2
Query: 168 QIHLQQDQSTALKCCHLPCWIAVSSTTSEIAS**RI*SWGPYGWNLWDWRLGKINTCESN 227
QIHLQQDQST L CCHLPCWIAVSSTTSEIAS** * WGPYGWNLWDWRL INTC+SN
Sbjct: 572 QIHLQQDQSTVLTCCHLPCWIAVSSTTSEIAS**GT**WGPYGWNLWDWRLR*INTCKSN 751
Query: 228 L*LYC*SI*MLMFS*KCEREFSFK*LEKSPAGAPFKNTST*N*VRKC*RRNS*NKGKATW 287
L* C*SI* MFS* +REFSFK*LE P APFK+TST N*V +C N +KGK
Sbjct: 752 L*FCC*SI*RFMFS*TSQREFSFK*LETFPGDAPFKDTSTEN*VSRCQ*GNLNHKGKVV* 931
Query: 288 KEDSFDS**C*QTGSARCFGWRT*LVRSWKQSHHHHPR*TLTRLSWDRKNICSRRVEWDR 347
KEDSFDS * *Q + CFGWR+*LVRSWK SHHHHPR*TLT LS DRKNICS+RVE DR
Sbjct: 932 KEDSFDSR*R*QYEATECFGWRS*LVRSWKPSHHHHPR*TLTSLS*DRKNICSKRVECDR 1111
Query: 348 SS*IVEVEGF*K*ESSFKL*RHFKACSCICFWPSISDRSSGF*LVWKEYCRM*VYIR*VW 407
SS*IVEV+GF*K*+SSFKL*+ F++CSC+CFWPS S R+SGF*LVWKE+ RM YIR V
Sbjct: 1112SS*IVEVDGF*K*QSSFKL*KDFESCSCLCFWPSGSYRNSGF*LVWKEHRRMQEYIRLV* 1291
Query: 408 KNST*RYPKDT*TQL*CFGRRGAECLFGYCLLHQRM*IGESQTDTSCSLWLFHRKPHWSV 467
K+S *R PK+T*++L* FG RGA+CL +CLL QR+*+G+ Q +TSCSLW H+ S
Sbjct: 1292KDSQ*RDPKNT*SEL*FFGGRGAKCLSRHCLLLQRL*MGKGQRNTSCSLWSLHKSSC*SA 1471
Query: 468 G**VSH-KY*LVLFQWYQSDIT*IDRGHG*RSRPTRIT*RAWRKKQVMVSR*YSSCFKRK 526
* +SH +*+ IT*+DR HG*R+RPTRIT AW+KKQVMV + Y +RK
Sbjct: 1472S*KMSH*PF*I----R*SCVIT*LDRKHG*RTRPTRITF*AWKKKQVMV*KGYI*SSRRK 1639
Query: 527 H 527
H
Sbjct: 1640H 1642
Score = 222 bits (566), Expect(2) = 0.0
Identities = 115/169 (68%), Positives = 128/169 (75%), Gaps = 2/169 (1%)
Frame = +3
Query: 1 MQSPSSSTSISYDYKYQVFLSFRGSDTRYGFTGNLYKALTDKGINTFIDDNG--LQRGNE 58
MQ P S+S SY + Y VFL +G+DTRYGFTGNL KAL DKGI TF DD+ LQR ++
Sbjct: 45 MQLPFYSSSFSYAFSYDVFLICKGTDTRYGFTGNLLKALIDKGIRTFHDDDDSDLQRRDK 224
Query: 59 ITPSLLKAIEESRIFIPVFSINYASSSFCLDELDHIIHCYKTKGRPVLPVFFGVDPSHVR 118
+TP + IEESRI IP+FS NYASSS CLD L HIIHCYKTKG VLPVFFGV+P+ VR
Sbjct: 225 VTPII---IEESRILIPIFSANYASSSSCLDTLVHIIHCYKTKGCLVLPVFFGVEPTDVR 395
Query: 119 HHKGSYGEALAEHEKRFQNDPKNMERLQGWKDALSQAANLSGYHDSPPG 167
HH G YG+ALAEHE RFQND KNMERLQ WK ALS AANL YHD G
Sbjct: 396 HHTGRYGKALAEHENRFQNDTKNMERLQQWKVALSLAANLPSYHDDSHG 542
>TC79023 weakly similar to GP|18033111|gb|AAL56987.1 functional candidate
resistance protein KR1 {Glycine max}, partial (37%)
Length = 2072
Score = 369 bits (946), Expect(2) = e-122
Identities = 216/361 (59%), Positives = 255/361 (69%)
Frame = +2
Query: 170 HLQQDQSTALKCCHLPCWIAVSSTTSEIAS**RI*SWGPYGWNLWDWRLGKINTCESNL* 229
H QQD S AL+CC + CWI V STT E AS**+I* GP+G LW WR INTC+SNL*
Sbjct: 725 HFQQD*S*ALRCCQISCWITVPSTTCERAS**KI***GPHGRTLWYWRHR*INTCKSNL* 904
Query: 230 LYC*SI*MLMFS*KCEREFSFK*LEKSPAGAPFKNTST*N*VRKC*RRNS*NKGKATWKE 289
+C*SI* MFS KCE EF F+*LE SP PFKN+ +*VR+C* NS + KA KE
Sbjct: 905 FHC*SI*SFMFSRKCESEFDFR*LETSPRETPFKNS*IGH*VRRC*SGNSNH*AKAM*KE 1084
Query: 290 DSFDS**C*QTGSARCFGWRT*LVRSWKQSHHHHPR*TLTRLSWDRKNICSRRVEWDRSS 349
DSFDS**C*QTGSAR GWRT*LVRSW+ SHH++ + TLT+ SWDRK CSRRVE DRSS
Sbjct: 1085 DSFDS**C*QTGSARGLGWRT*LVRSWE*SHHYYSKQTLTKDSWDRKYTCSRRVECDRSS 1264
Query: 350 *IVEVEGF*K*ESSFKL*RHFKACSCICFWPSISDRSSGF*LVWKEYCRM*VYIR*VWKN 409
IVEV+GF + + SFK *RHFK C+ +CFWPSIS ++ WKE R YIR V +N
Sbjct: 1265 RIVEVDGFQR-KCSFKS*RHFKPCTYLCFWPSISYCNNRVQFGWKERTRFNEYIRWV*RN 1441
Query: 410 ST*RYPKDT*TQL*CFGRRGAECLFGYCLLHQRM*IGESQTDTSCSLWLFHRKPHWSVG* 469
S *R PK+T*++L* FG R A+CL +CLL QR+*+ Q +TSCSLW H S G*
Sbjct: 1442 SQ*RDPKNT*SEL*FFGERRAKCLSRHCLLLQRV*MARGQRNTSCSLWSLHSTSCCSFG* 1621
Query: 470 *VSHKY*LVLFQWYQSDIT*IDRGHG*RSRPTRIT*RAWRKKQVMVSR*YSSCFKRKHYK 529
+SH + DIT*+DRGHG*RSRPTRIT AWR + V+V *YSSCFK+KH+
Sbjct: 1622 KISHGPFEI---**LCDIT*LDRGHG*RSRPTRITR*AWRTQ*VVV*A*YSSCFKKKHWN 1792
Query: 530 K 530
K
Sbjct: 1793 K 1795
Score = 262 bits (669), Expect = 4e-70
Identities = 141/207 (68%), Positives = 153/207 (73%), Gaps = 9/207 (4%)
Frame = +1
Query: 2 QSPSSSTSISYDYKYQVFLSFRGSDTRYGFTGNLYKALTDKGINTFIDDNGLQRGNEITP 61
QSPSS T YQVFLSFRG+DTR+GFTGNLYKALTDKGI TFIDDN LQRG+EITP
Sbjct: 73 QSPSSFT-------YQVFLSFRGADTRHGFTGNLYKALTDKGIYTFIDDNDLQRGDEITP 231
Query: 62 SLLKAIEESRIFIPVFSINYASSSFCLDELDHIIHCYKTKGRPVLPVFFGVDPSHVRHHK 121
SL AIE+SRIFIPVFS NYASSSFCLDEL HI HCY TKG VLPVF GVDP+ VRHH
Sbjct: 232 SLKNAIEKSRIFIPVFSENYASSSFCLDELVHITHCYDTKGCLVLPVFIGVDPTDVRHHT 411
Query: 122 GSYGEALAEHEKRFQNDPKNMERLQGWKDALSQAANLSGYHDSPPGQI---------HLQ 172
G YGEALA H+K+FQND N ERLQ WK+ALSQAANLSG H +I H Q
Sbjct: 412 GRYGEALAVHKKKFQNDKDNTERLQQWKEALSQAANLSGQHYKHGYRIRVYWKDSRRHFQ 591
Query: 173 QDQSTALKCCHLPCWIAVSSTTSEIAS 199
QD S A +C +PC I STT + AS
Sbjct: 592 QD*SQAPRCSQIPC*ITAPSTTYKNAS 672
Score = 87.8 bits (216), Expect(2) = e-122
Identities = 51/98 (52%), Positives = 60/98 (61%)
Frame = +3
Query: 531 GKAFKKMTRLKTLIIENGHCSKGLKYLRSSLKALKWEGCLSKSLSSSILSKASEIKSFSN 590
G AF+KMT LKT I ENGH SK L+YL SSL+ +K GC+ KS SSS
Sbjct: 1854 GNAFEKMTNLKTFITENGHHSKSLEYLPSSLRVMK--GCIPKSPSSSS------------ 1991
Query: 591 CIYL*CTMISFSAKKFQDMTILILDHCEYLTHIPDVSG 628
S KKF+DM +LIL++CEYLTHIPDVSG
Sbjct: 1992 -----------SNKKFEDMKVLILNNCEYLTHIPDVSG 2072
>TC92115 weakly similar to GP|18033111|gb|AAL56987.1 functional candidate
resistance protein KR1 {Glycine max}, partial (24%)
Length = 1348
Score = 292 bits (748), Expect(2) = e-103
Identities = 166/274 (60%), Positives = 200/274 (72%), Gaps = 1/274 (0%)
Frame = +1
Query: 256 SPAGAPFKNTST*N*VRKC*RRNS*NKGKATWKEDSFDS**C*QTGSARCFGWRT*LVRS 315
SP GAPFK+TST N*VR+C* NS KGKA +KEDSFDS *C*+ G+ CFG RT*LVR
Sbjct: 1 SPGGAPFKSTSTEN*VRRC**GNSLYKGKAAYKEDSFDSR*C*RYGATTCFGRRT*LVRP 180
Query: 316 WKQSHHHHPR*TLTRLSWDRKNICSRRVEWDRSS*IVEVEGF*K*ESSFKL*RHFKACSC 375
WKQ HH+H R TLT+ SWDRK S R WDRSS +VE++GF*+*+S+F L*R FK+CS
Sbjct: 181 WKQGHHYHSRQTLTKKSWDRKYA*SGRAVWDRSSRVVEMDGF*E*QSTFNL*RRFKSCSF 360
Query: 376 ICFWPSISDRSSGF*LVWKEYCRM*VYIR*VWKNST*RYPKDT*TQL*CFGRRGAECLFG 435
ICF PS+S R+SGF LVWK+ RM YIR + K+S *+ P DT*++L*CFG R A+C
Sbjct: 361 ICFGPSLSFRNSGFQLVWKDDRRMEGYIRWI*KDSQ*KDPPDT*SKL*CFGGRAAKCFSR 540
Query: 436 YCLLHQRM*IGESQTDTSCSLWLFHRKPHWSVG**VSH-KY*LVLFQWYQSDIT*IDRGH 494
+CLL QRM +G TSCSLW H SVG* +S Y L+ + +D T*+D+G+
Sbjct: 541 HCLLLQRMWMGGV*IYTSCSLWPSHNTSSSSVG*KISR*DYSSPLWIYL*TDTT*LDKGN 720
Query: 495 G*RSRPTRIT*RAWRKKQVMVSR*YSSCFKRKHY 528
G*RSRPTRI RAWRKK +MV R*YS CFKRKH+
Sbjct: 721 G*RSRPTRIAQRAWRKK*IMV*R*YS*CFKRKHW 822
Score = 103 bits (257), Expect(2) = e-103
Identities = 69/143 (48%), Positives = 79/143 (54%), Gaps = 26/143 (18%)
Frame = +2
Query: 529 KKGKAFKKMTRLKTLIIENGHCSKGLKYLRSSLKALKWEGCLSKSLSSSILSKASEIKSF 588
KKGKAFKKMTRLKTLIIEN H SKGLKYL SSL+ LK GCLS+SL S LSKA EI SF
Sbjct: 881 KKGKAFKKMTRLKTLIIENVHFSKGLKYLPSSLRVLKLRGCLSESLISCSLSKAIEITSF 1060
Query: 589 SNCIYL*C-TMISFSAKKFQDMTILILDHCEYLTHIPDV--------------------- 626
S CIYL F ++ ++ + +P V
Sbjct: 1061 SYCIYLLIYNDFCFRQQRRNRYDSIVFNF*LIYLFLPXVYAEVPEHENLDIGQV*IF*PI 1240
Query: 627 ----SGLSNLEKLSFEYCKNLIT 645
SGL NL+K SFEY +NLIT
Sbjct: 1241 YLMXSGLQNLKKFSFEYXENLIT 1309
Score = 36.2 bits (82), Expect = 0.048
Identities = 24/63 (38%), Positives = 32/63 (50%), Gaps = 1/63 (1%)
Frame = +3
Query: 596 CTMISFSAKKFQDMTILILDHCEYLTHIPDVSGLSNLEKLS-FEYCKNLITIHNSIGHLN 654
C + F +KFQ+M IL LD CEY + + K S + + IHNSIGHL
Sbjct: 1161 CFFLXFM-QKFQNMKILTLDRCEYFNPYT*CXQVFKI*KSSPLNIXRI*LPIHNSIGHLI 1337
Query: 655 KLE 657
L+
Sbjct: 1338 NLK 1346
>BF651081 weakly similar to GP|18033111|g functional candidate resistance
protein KR1 {Glycine max}, partial (16%)
Length = 664
Score = 251 bits (641), Expect(3) = 4e-99
Identities = 122/138 (88%), Positives = 130/138 (93%)
Frame = +1
Query: 30 GFTGNLYKALTDKGINTFIDDNGLQRGNEITPSLLKAIEESRIFIPVFSINYASSSFCLD 89
GFTGNLYKAL DKGI+TFID++ LQRG+EITPSLLKAIEESRIFI VFSINYASSSFCLD
Sbjct: 67 GFTGNLYKALDDKGIHTFIDNHELQRGDEITPSLLKAIEESRIFIAVFSINYASSSFCLD 246
Query: 90 ELDHIIHCYKTKGRPVLPVFFGVDPSHVRHHKGSYGEALAEHEKRFQNDPKNMERLQGWK 149
EL HIIHCYKTKGR VLPVFF V+P+ VRH KGSYGEALAEHEKRFQNDPK+MERLQGWK
Sbjct: 247 ELVHIIHCYKTKGRLVLPVFFAVEPTIVRHQKGSYGEALAEHEKRFQNDPKSMERLQGWK 426
Query: 150 DALSQAANLSGYHDSPPG 167
+ALSQAANLSGYHDSPPG
Sbjct: 427 EALSQAANLSGYHDSPPG 480
Score = 104 bits (260), Expect(3) = 4e-99
Identities = 46/51 (90%), Positives = 47/51 (91%)
Frame = +3
Query: 168 QIHLQQDQSTALKCCHLPCWIAVSSTTSEIAS**RI*SWGPYGWNLWDWRL 218
QIHLQQDQST L CCHLPCWIAVS TT+EIAS** I*SWGPYGWNLWDWRL
Sbjct: 510 QIHLQQDQSTTLTCCHLPCWIAVSXTTNEIAS**XI*SWGPYGWNLWDWRL 662
Score = 46.6 bits (109), Expect(3) = 4e-99
Identities = 20/23 (86%), Positives = 22/23 (94%)
Frame = +3
Query: 9 SISYDYKYQVFLSFRGSDTRYGF 31
SISY+YKYQVFL+FRGSDTRY F
Sbjct: 3 SISYEYKYQVFLNFRGSDTRYXF 71
>TC77731 weakly similar to GP|18033111|gb|AAL56987.1 functional candidate
resistance protein KR1 {Glycine max}, partial (23%)
Length = 1651
Score = 339 bits (870), Expect = 2e-93
Identities = 195/326 (59%), Positives = 230/326 (69%)
Frame = +3
Query: 170 HLQQDQSTALKCCHLPCWIAVSSTTSEIAS**RI*SWGPYGWNLWDWRLGKINTCESNL* 229
HLQQ QS K +PCWI V + TSEIAS* +* GP+G W WR G INTC+S+L*
Sbjct: 534 HLQQYQSCFFKRSQIPCWITVPNRTSEIAS*HGL*G*GPHGRTFWYWRHG*INTCKSSL* 713
Query: 230 LYC*SI*MLMFS*KCEREFSFK*LEKSPAGAPFKNTST*N*VRKC*RRNS*NKGKATWKE 289
C*SI* MFS C+REF+ E SP APFKN+ * *VR+C* N+ +KGK KE
Sbjct: 714 FCC*SI*RCMFSS*CQREFNS*EPETSPKEAPFKNS*I*R*VRRC**GNTNHKGKTI*KE 893
Query: 290 DSFDS**C*QTGSARCFGWRT*LVRSWKQSHHHHPR*TLTRLSWDRKNICSRRVEWDRSS 349
DSFDS**C*QTG+AR FGWRT*LVRSWKQ HH+H *TLT LSWD K CSRR+E DRSS
Sbjct: 894 DSFDS**C*QTGAARGFGWRT*LVRSWKQGHHYHS**TLTSLSWDYKYTCSRRIE*DRSS 1073
Query: 350 *IVEVEGF*K*ESSFKL*RHFKACSCICFWPSISDRSSGF*LVWKEYCRM*VYIR*VWKN 409
*IVE GF*K*+SSF L*R FK CS +C W ++S+ ++ * VWKE R+ YIR*V K
Sbjct: 1074*IVEANGF*K*QSSFNL*RDFKPCSYLCIWAALSNSNNRG*FVWKEGRRLEAYIR*V*KY 1253
Query: 410 ST*RYPKDT*TQL*CFGRRGAECLFGYCLLHQRM*IGESQTDTSCSLWLFHRKPHWSVG* 469
S *RYPKDT + L*CFG +G ECL +CLL QR+*+ + +TSCSLW HR WSVG*
Sbjct: 1254SQ*RYPKDTSS*L*CFGTKGEECLSRHCLLLQRV*MDKG*KNTSCSLWPLHRTSCWSVG* 1433
Query: 470 *VSHKY*LVLFQWYQSDIT*IDRGHG 495
+SH+ L + D+T*+DRGHG
Sbjct: 1434KISHR---PLGI*HSDDLT*LDRGHG 1502
Score = 137 bits (346), Expect = 1e-32
Identities = 64/87 (73%), Positives = 73/87 (83%)
Frame = +2
Query: 81 YASSSFCLDELDHIIHCYKTKGRPVLPVFFGVDPSHVRHHKGSYGEALAEHEKRFQNDPK 140
YASSSFCLDEL HIIHCYKTK VLPVF+ V+P+H+RH GSYGE L +HE+RFQN+ K
Sbjct: 2 YASSSFCLDELVHIIHCYKTKSCLVLPVFYDVEPTHIRHQSGSYGEYLTKHEERFQNNEK 181
Query: 141 NMERLQGWKDALSQAANLSGYHDSPPG 167
NMERL+ WK AL+QAANLSGYH SP G
Sbjct: 182 NMERLRQWKIALTQAANLSGYHYSPHG 262
Score = 37.4 bits (85), Expect = 0.021
Identities = 29/77 (37%), Positives = 36/77 (46%)
Frame = +3
Query: 552 KGLKYLRSSLKALKWEGCLSKSLSSSILSKASEIKSFSNCIYL*CTMISFSAKKFQDMTI 611
+G YL SSL+ L+W KSLS CI +K+F M +
Sbjct: 1491 RGHGYLPSSLRYLEWIDYDFKSLS---------------CIL---------SKEFNYMKV 1598
Query: 612 LILDHCEYLTHIPDVSG 628
L LD+ LTHIPDVSG
Sbjct: 1599 LKLDYSSDLTHIPDVSG 1649
>TC80083 weakly similar to GP|18033111|gb|AAL56987.1 functional candidate
resistance protein KR1 {Glycine max}, partial (4%)
Length = 1269
Score = 305 bits (781), Expect = 4e-83
Identities = 160/221 (72%), Positives = 173/221 (77%)
Frame = +3
Query: 529 KKGKAFKKMTRLKTLIIENGHCSKGLKYLRSSLKALKWEGCLSKSLSSSILSKASEIKSF 588
+KGKAFKKMT+LKTLIIENGHCSKGLKYL SSL LKW+GCLSK LSS IL+K
Sbjct: 78 EKGKAFKKMTKLKTLIIENGHCSKGLKYLPSSLSVLKWKGCLSKCLSSRILNK------- 236
Query: 589 SNCIYL*CTMISFSAKKFQDMTILILDHCEYLTHIPDVSGLSNLEKLSFEYCKNLITIHN 648
KFQDM +LILD C+YLTHIPDVSGLSNLEKLSFE C NLITIHN
Sbjct: 237 ----------------KFQDMKVLILDRCKYLTHIPDVSGLSNLEKLSFERCYNLITIHN 368
Query: 649 SIGHLNKLERLSAFGCRTLKRFPPLGLASLKELKLSCCYSLKSFPKLLCKMTNIDKIWFW 708
SIGHLNKLERLSA+GC ++ FPPLGLASLKEL LS C SLKSFP+LLCKMTNID IW
Sbjct: 369 SIGHLNKLERLSAYGCNKVEHFPPLGLASLKELNLSSCRSLKSFPELLCKMTNIDNIWLC 548
Query: 709 YTSIRELPSSFQNLSELDELSVREFGMLRFPKHNDRMYSIV 749
TSI ELP SFQNLSEL +LSV +GMLRFPK ND+MYSIV
Sbjct: 549 NTSIGELPFSFQNLSELHKLSV-TYGMLRFPKQNDKMYSIV 668
>TC84226 similar to GP|22037313|gb|AAM89998.1 disease resistance-like
protein GS0-1 {Glycine max}, partial (72%)
Length = 665
Score = 264 bits (674), Expect(2) = 3e-79
Identities = 131/159 (82%), Positives = 143/159 (89%)
Frame = +3
Query: 7 STSISYDYKYQVFLSFRGSDTRYGFTGNLYKALTDKGINTFIDDNGLQRGNEITPSLLKA 66
S+SISY +KYQVFLSFRGSDTRYGFTGNLYKALTDKGI+TFIDD+ LQRG+EITPSL A
Sbjct: 3 SSSISYVFKYQVFLSFRGSDTRYGFTGNLYKALTDKGIHTFIDDSELQRGDEITPSLDNA 182
Query: 67 IEESRIFIPVFSINYASSSFCLDELDHIIHCYKTKGRPVLPVFFGVDPSHVRHHKGSYGE 126
IEESRIFIPVFS NYASSSFCLDEL HIIH YK GR VLPVFFGVDPSHVRHH+GSYGE
Sbjct: 183 IEESRIFIPVFSANYASSSFCLDELVHIIHLYKQNGRLVLPVFFGVDPSHVRHHRGSYGE 362
Query: 127 ALAEHEKRFQNDPKNMERLQGWKDALSQAANLSGYHDSP 165
ALA+HE+RFQ++ +MERLQ WK AL+QAANLSG H SP
Sbjct: 363 ALAKHEERFQHNTDHMERLQKWKIALTQAANLSGDHRSP 479
Score = 50.8 bits (120), Expect(2) = 3e-79
Identities = 22/27 (81%), Positives = 23/27 (84%)
Frame = +2
Query: 168 QIHLQQDQSTALKCCHLPCWIAVSSTT 194
QIHLQQDQS AL CC LPCWI VS+TT
Sbjct: 584 QIHLQQDQSCALTCCQLPCWI*VSNTT 664
>TC77730 weakly similar to GP|7341113|gb|AAF61210.1| unknown {Glycine max},
partial (80%)
Length = 831
Score = 232 bits (592), Expect(3) = 5e-78
Identities = 119/165 (72%), Positives = 132/165 (79%)
Frame = +1
Query: 1 MQSPSSSTSISYDYKYQVFLSFRGSDTRYGFTGNLYKALTDKGINTFIDDNGLQRGNEIT 60
MQSPS +VFL+FRGSDTR FTGNLYKAL DKGI TFID+N LQRG+EIT
Sbjct: 22 MQSPS-----------RVFLNFRGSDTRNNFTGNLYKALVDKGIRTFIDENDLQRGDEIT 168
Query: 61 PSLLKAIEESRIFIPVFSINYASSSFCLDELDHIIHCYKTKGRPVLPVFFGVDPSHVRHH 120
PSL+KAIEESRI IP+FS NYASSSFCLDEL HIIHCYKTK VLPVF+GVDP+ VR H
Sbjct: 169 PSLVKAIEESRICIPIFSANYASSSFCLDELVHIIHCYKTKSCLVLPVFYGVDPTDVRRH 348
Query: 121 KGSYGEALAEHEKRFQNDPKNMERLQGWKDALSQAANLSGYHDSP 165
SYGE L +HE+ FQN+ KNMERL+ WK AL+QAANLSGYH SP
Sbjct: 349 TCSYGEYLTKHEEGFQNNEKNMERLRQWKMALTQAANLSGYHYSP 483
Score = 70.1 bits (170), Expect(3) = 5e-78
Identities = 40/72 (55%), Positives = 46/72 (63%)
Frame = +3
Query: 163 DSPPGQIHLQQDQSTALKCCHLPCWIAVSSTTSEIAS**RI*SWGPYGWNLWDWRLGKIN 222
DSP IHLQQ QS K +PCWI V + TSEIAS* +* GP+G W WR G IN
Sbjct: 513 DSP---IHLQQYQS*FFKRSQIPCWITVPNRTSEIAS*HGL*G*GPHGRTFWYWRHG*IN 683
Query: 223 TCESNL*LYC*S 234
TC+S+ * C*S
Sbjct: 684 TCKSSF*FSC*S 719
Score = 29.3 bits (64), Expect(3) = 5e-78
Identities = 17/29 (58%), Positives = 18/29 (61%)
Frame = +2
Query: 235 I*MLMFS*KCEREFSFK*LEKSPAGAPFK 263
I* MFS C+REF * E SP APFK
Sbjct: 719 I*RCMFSS*CQREFKS**PETSPKDAPFK 805
>TC92835 weakly similar to GP|18033111|gb|AAL56987.1 functional candidate
resistance protein KR1 {Glycine max}, partial (13%)
Length = 581
Score = 278 bits (712), Expect(2) = 3e-76
Identities = 137/165 (83%), Positives = 146/165 (88%)
Frame = +2
Query: 1 MQSPSSSTSISYDYKYQVFLSFRGSDTRYGFTGNLYKALTDKGINTFIDDNGLQRGNEIT 60
MQ PSSS+S S + YQVFLSFRG+DTR+GFTGNLYKALTDKGI TFIDDN LQRG+EIT
Sbjct: 17 MQLPSSSSSFSSAFTYQVFLSFRGTDTRHGFTGNLYKALTDKGIKTFIDDNDLQRGDEIT 196
Query: 61 PSLLKAIEESRIFIPVFSINYASSSFCLDELDHIIHCYKTKGRPVLPVFFGVDPSHVRHH 120
PSLLKAIEESRIFIPVFSINYA+S FCLDEL HIIHCYKTKGR VLPVFFGVDP++VRHH
Sbjct: 197 PSLLKAIEESRIFIPVFSINYATSKFCLDELVHIIHCYKTKGRLVLPVFFGVDPTNVRHH 376
Query: 121 KGSYGEALAEHEKRFQNDPKNMERLQGWKDALSQAANLSGYHDSP 165
G YGEALA HEKRFQND NMERL WK AL+QAANLSGYH SP
Sbjct: 377 TGPYGEALAGHEKRFQNDKDNMERLHQWKLALTQAANLSGYHSSP 511
Score = 25.8 bits (55), Expect(2) = 3e-76
Identities = 11/12 (91%), Positives = 12/12 (99%)
Frame = +1
Query: 168 QIHLQQDQSTAL 179
QIHLQQDQS+AL
Sbjct: 544 QIHLQQDQSSAL 579
>TC77728 similar to GP|12056928|gb|AAG48132.1 putative resistance protein
{Glycine max}, partial (12%)
Length = 765
Score = 235 bits (600), Expect(2) = 8e-76
Identities = 119/167 (71%), Positives = 133/167 (79%)
Frame = +1
Query: 1 MQSPSSSTSISYDYKYQVFLSFRGSDTRYGFTGNLYKALTDKGINTFIDDNGLQRGNEIT 60
MQSPS +VFLSFRGSDTR FTGNLYKAL DKGI TFIDDN L+RG+EIT
Sbjct: 76 MQSPS-----------RVFLSFRGSDTRNKFTGNLYKALVDKGIRTFIDDNDLERGDEIT 222
Query: 61 PSLLKAIEESRIFIPVFSINYASSSFCLDELDHIIHCYKTKGRPVLPVFFGVDPSHVRHH 120
PSL+KAIEESRIFIP+FS NYASSSFCLDEL HIIHCYKTK V PVF+ V+P+H+R+
Sbjct: 223 PSLVKAIEESRIFIPIFSANYASSSFCLDELVHIIHCYKTKSCLVFPVFYDVEPTHIRNQ 402
Query: 121 KGSYGEALAEHEKRFQNDPKNMERLQGWKDALSQAANLSGYHDSPPG 167
G YGE L +HE+RFQN+ KNMERL+ WK AL QAANLSGYH SP G
Sbjct: 403 SGIYGEHLTKHEERFQNNEKNMERLRQWKIALIQAANLSGYHYSPHG 543
Score = 67.8 bits (164), Expect(2) = 8e-76
Identities = 34/60 (56%), Positives = 40/60 (66%)
Frame = +3
Query: 170 HLQQDQSTALKCCHLPCWIAVSSTTSEIAS**RI*SWGPYGWNLWDWRLGKINTCESNL* 229
HLQQ QS KC +PCWI V + SEIAS* +* GP+G W WR G INTC+S+L*
Sbjct: 579 HLQQYQSCFFKCSQIPCWITVPNRRSEIAS*HGL*G*GPHGRTFWYWRHG*INTCKSSL* 758
>TC86227 weakly similar to GP|18033111|gb|AAL56987.1 functional candidate
resistance protein KR1 {Glycine max}, partial (22%)
Length = 1736
Score = 276 bits (705), Expect = 3e-74
Identities = 162/250 (64%), Positives = 186/250 (73%)
Frame = +3
Query: 170 HLQQDQSTALKCCHLPCWIAVSSTTSEIAS**RI*SWGPYGWNLWDWRLGKINTCESNL* 229
H + DQS C +PCWI V STTS+IAS * I* G +G LW LGKINTC+SNL*
Sbjct: 717 HFR*DQSCFSTRCQIPCWITVPSTTSKIASG*GI**RGQHGRTLWY*SLGKINTCKSNL* 896
Query: 230 LYC*SI*MLMFS*KCEREFSFK*LEKSPAGAPFKNTST*N*VRKC*RRNS*NKGKATWKE 289
+C*SI* M S C REFS LE SP GAPFKN+ST +*V++C NS + GKA +E
Sbjct: 897 FHC*SI*RSMLSS*C*REFSS*KLETSPEGAPFKNSSTEH*VKRCF*GNSNH*GKAM*EE 1076
Query: 290 DSFDS**C*QTGSARCFGWRT*LVRSWKQSHHHHPR*TLTRLSWDRKNICSRRVEWDRSS 349
DSFDS *C* TGSAR FGWRT*LVRSWKQSHHHHPR*TLT LSWDRKNICS+R W+ SS
Sbjct: 1077DSFDSR*C*PTGSARGFGWRT*LVRSWKQSHHHHPR*TLTNLSWDRKNICSKRAIWEGSS 1256
Query: 350 *IVEVEGF*K*ESSFKL*RHFKACSCICFWPSISDRSSGF*LVWKEYCRM*VYIR*VWKN 409
*IVEV+GF*+*+S+ KL*R FK CS I F S+S R+SGF*LVWKE+ M +IR V ++
Sbjct: 1257*IVEVDGF*E*QSTAKL*RCFKPCSFIWFGHSLSFRNSGF*LVWKEHRSMEEHIRWV*QD 1436
Query: 410 ST*RYPKDT* 419
S *R KDT*
Sbjct: 1437SQ*RDTKDT* 1466
Score = 235 bits (599), Expect = 5e-62
Identities = 113/154 (73%), Positives = 128/154 (82%)
Frame = +3
Query: 11 SYDYKYQVFLSFRGSDTRYGFTGNLYKALTDKGINTFIDDNGLQRGNEITPSLLKAIEES 70
S +KY VFLSFR DT YGFTGNLYKAL DKGI TFIDDN L+RG+E TPSL+KAIEES
Sbjct: 3 SSGFKYHVFLSFRDIDTLYGFTGNLYKALIDKGIKTFIDDNDLERGDESTPSLVKAIEES 182
Query: 71 RIFIPVFSINYASSSFCLDELDHIIHCYKTKGRPVLPVFFGVDPSHVRHHKGSYGEALAE 130
RI IP+FS NYASSSFCLDEL HIIHCYKT+G VLPVF+G DP+HVRH GSYGE L +
Sbjct: 183 RILIPIFSANYASSSFCLDELVHIIHCYKTRGCSVLPVFYGADPTHVRHQTGSYGEHLTK 362
Query: 131 HEKRFQNDPKNMERLQGWKDALSQAANLSGYHDS 164
HE +FQN+ +NMERL+ WK AL+QAAN SG+H S
Sbjct: 363 HEDKFQNNKENMERLKKWKMALTQAANFSGHHFS 464
>BF646053 weakly similar to GP|18033111|g functional candidate resistance
protein KR1 {Glycine max}, partial (9%)
Length = 644
Score = 275 bits (702), Expect = 6e-74
Identities = 149/191 (78%), Positives = 152/191 (79%), Gaps = 1/191 (0%)
Frame = +3
Query: 560 SLKALKWEGCLSKSLSSSILSKASEIKSFSNCIYL*CTMISFSAKKFQDMTILILDHCEY 619
SLKALKWEGCLSKSLSSSILSK KFQDMTILILDHCEY
Sbjct: 3 SLKALKWEGCLSKSLSSSILSK-----------------------KFQDMTILILDHCEY 113
Query: 620 LTHIPDVSGLSNLEKLSFEYCKNLITIHNSIGHLNKLERLSAFGCRTLKRFPPLGLASLK 679
LTHIPDVSGLSNLEKLSFE C NLITIHNSIGHLNKLERLSAFGCR LKRFPPLGLASLK
Sbjct: 114 LTHIPDVSGLSNLEKLSFECCYNLITIHNSIGHLNKLERLSAFGCRKLKRFPPLGLASLK 293
Query: 680 ELKLSCCYSLKSFPKLLCKMTNIDKIWFWYT-SIRELPSSFQNLSELDELSVREFGMLRF 738
EL + CC SLKSFP+LLCKMTNI +I Y SI ELPSSFQNLSELDELSVRE MLRF
Sbjct: 294 ELDICCCSSLKSFPELLCKMTNIKEIDLDYNISIGELPSSFQNLSELDELSVREARMLRF 473
Query: 739 PKHNDRMYSIV 749
PKHNDRMYS V
Sbjct: 474 PKHNDRMYSKV 506
>TC86225 weakly similar to GP|9858476|gb|AAG01051.1| resistance protein LM12
{Glycine max}, partial (25%)
Length = 677
Score = 247 bits (631), Expect(2) = 5e-69
Identities = 121/164 (73%), Positives = 137/164 (82%)
Frame = +3
Query: 1 MQSPSSSTSISYDYKYQVFLSFRGSDTRYGFTGNLYKALTDKGINTFIDDNGLQRGNEIT 60
MQS SSS+SIS +KY VFLSFR DT YGFTGNLYKAL DKGI TFIDDN L+RG+E T
Sbjct: 66 MQSLSSSSSISSGFKYHVFLSFRDIDTLYGFTGNLYKALIDKGIKTFIDDNDLERGDEST 245
Query: 61 PSLLKAIEESRIFIPVFSINYASSSFCLDELDHIIHCYKTKGRPVLPVFFGVDPSHVRHH 120
PSL+KAIEESRI IP+FS NYASSSFCLDEL HIIHCYKT+G VLPVF+G DP+HVRH
Sbjct: 246 PSLVKAIEESRILIPIFSANYASSSFCLDELVHIIHCYKTRGCSVLPVFYGADPTHVRHQ 425
Query: 121 KGSYGEALAEHEKRFQNDPKNMERLQGWKDALSQAANLSGYHDS 164
GSYGE L +HE +FQN+ +NMERL+ WK AL+QAAN SG+H S
Sbjct: 426 TGSYGEHLTKHEDKFQNNKENMERLKKWKMALTQAANFSGHHFS 557
Score = 33.1 bits (74), Expect(2) = 5e-69
Identities = 14/26 (53%), Positives = 16/26 (60%)
Frame = +2
Query: 170 HLQQDQSTALKCCHLPCWIAVSSTTS 195
H + DQS C +PCWI V STTS
Sbjct: 599 HFR*DQSCFSTRCQIPCWITVPSTTS 676
>AW688466 similar to GP|9965109|gb| resistance protein MG13 {Glycine max},
partial (31%)
Length = 668
Score = 250 bits (638), Expect = 2e-66
Identities = 128/207 (61%), Positives = 151/207 (72%), Gaps = 11/207 (5%)
Frame = +2
Query: 1 MQSPSSSTSISYDYKYQVFLSFRGSDTRYGFTGNLYKALTDKGINTFIDDNGLQRGNEIT 60
MQSPSSS+S+SY + + VF+SFRG+DTRYGFTGNLYKAL+DKGI TFIDD LQRG+EIT
Sbjct: 41 MQSPSSSSSLSYGFSFDVFISFRGTDTRYGFTGNLYKALSDKGIRTFIDDKELQRGDEIT 220
Query: 61 PSLLKAIEESRIFIPVFSINYASSSFCLDELDHIIHCYKTKGRPVLPVFFGVDPSHVRHH 120
PSLLK+IE+SRI I VFS +YASSSFCL EL HII C KG V+PVF+G +PS VRH
Sbjct: 221 PSLLKSIEDSRIAIIVFSKDYASSSFCLXELVHIIQCSNEKGTTVIPVFYGTEPSQVRHQ 400
Query: 121 KGSYGEALAEHEKRFQNDPKNMERLQGWKDALSQAANLSGYHDSPPGQI----------- 169
SYGEALA+HE+ FQN +NMERL WK AL+QAANLSG+H I
Sbjct: 401 NDSYGEALAKHEEGFQNSKENMERLLKWKKALNQAANLSGHHFQSRK*I*T*FY*KNXYR 580
Query: 170 HLQQDQSTALKCCHLPCWIAVSSTTSE 196
H QQD ++ CC LP W +S+ SE
Sbjct: 581 HXQQD*PRSITCCRLPYWSQISNIRSE 661
>TC77733 weakly similar to GP|18033111|gb|AAL56987.1 functional candidate
resistance protein KR1 {Glycine max}, partial (12%)
Length = 632
Score = 244 bits (623), Expect = 9e-65
Identities = 122/169 (72%), Positives = 137/169 (80%)
Frame = +3
Query: 1 MQSPSSSTSISYDYKYQVFLSFRGSDTRYGFTGNLYKALTDKGINTFIDDNGLQRGNEIT 60
MQSPS +VFLSFRGSDTR FTGNLYKAL DKGI TFIDDN L+RG+EIT
Sbjct: 36 MQSPS-----------RVFLSFRGSDTRNNFTGNLYKALVDKGIRTFIDDNDLERGDEIT 182
Query: 61 PSLLKAIEESRIFIPVFSINYASSSFCLDELDHIIHCYKTKGRPVLPVFFGVDPSHVRHH 120
PSL+KAIEESRIFIP+FS NYASSSFCLDEL HIIHCYKTK VLPVF+ V+P+H+RH
Sbjct: 183 PSLVKAIEESRIFIPIFSANYASSSFCLDELVHIIHCYKTKSCLVLPVFYDVEPTHIRHQ 362
Query: 121 KGSYGEALAEHEKRFQNDPKNMERLQGWKDALSQAANLSGYHDSPPGQI 169
GSYGE L +HE+RFQN+ KNMERL+ WK AL QAANLSGYH SP G++
Sbjct: 363 SGSYGEHLTKHEERFQNNEKNMERLRQWKIALIQAANLSGYHYSPHGEL 509
>BF646616 similar to GP|9965103|gb|A resistance protein LM6 {Glycine max},
partial (5%)
Length = 671
Score = 238 bits (608), Expect = 5e-63
Identities = 133/220 (60%), Positives = 152/220 (68%), Gaps = 1/220 (0%)
Frame = +2
Query: 531 GKAFKKMTRLKTLIIENGHCSKGLKYLRSSLKALKWEGCLSKSLSSSILSKASEIKSFSN 590
G AFKKMT LK LIIENGH S+G YL SSL+ KWE SKSLS
Sbjct: 8 GNAFKKMTNLKILIIENGHFSEGPNYLPSSLRCWKWEEYPSKSLSC-------------- 145
Query: 591 CIYL*CTMISFSAKKFQDMTILILDHCEYLTHIPDVSGLSNLEKLSFEYCKNLITIHNSI 650
FS KKF+++ +LILD C+YLT I +VS L NLE+ SF+ CKNLITIHNSI
Sbjct: 146 ----------FSNKKFENLKVLILDRCQYLTDISNVSNLPNLEEFSFQDCKNLITIHNSI 295
Query: 651 GHLNKLERLSAFGCRTLKRFPPLGLASLKELKLSCCYSLKSFPKLLCKMTNIDKIW-FWY 709
GHLNKLE + A GC L+ FPPL L SLKEL+LS C SLKSFP+LLCKMTNI I+ F++
Sbjct: 296 GHLNKLEIIRATGCCKLESFPPLLLPSLKELELSYCESLKSFPELLCKMTNIKDIFLFFH 475
Query: 710 TSIRELPSSFQNLSELDELSVREFGMLRFPKHNDRMYSIV 749
TSI ELP SFQNLSEL L + MLRFPKHND+MYSIV
Sbjct: 476 TSIEELPFSFQNLSELHVLQIYHVRMLRFPKHNDKMYSIV 595
>BF650880 weakly similar to GP|9858476|gb| resistance protein LM12 {Glycine
max}, partial (41%)
Length = 659
Score = 235 bits (600), Expect(2) = 4e-62
Identities = 130/191 (68%), Positives = 140/191 (73%)
Frame = +2
Query: 168 QIHLQQDQSTALKCCHLPCWIAVSSTTSEIAS**RI*SWGPYGWNLWDWRLGKINTCESN 227
QIHLQQDQS+AL CC LPCWIA+SST SEIAS** * WGPYGWNLWDWRLG I+TC+SN
Sbjct: 56 QIHLQQDQSSALTCC*LPCWIALSSTRSEIAS**GT**WGPYGWNLWDWRLG*ISTCKSN 235
Query: 228 L*LYC*SI*MLMFS*KCEREFSFK*LEKSPAGAPFKNTST*N*VRKC*RRNS*NKGKATW 287
L* C*SI* MFS CEREFS K*LE SP A KN N VR C +S KGK
Sbjct: 236 L*FCC*SI*RFMFSS*CEREFSSK*LETSPREASSKNNWIENQVRSCL*GDSNYKGKTV* 415
Query: 288 KEDSFDS**C*QTGSARCFGWRT*LVRSWKQSHHHHPR*TLTRLSWDRKNICSRRVEWDR 347
K DSFDS *C*+ G+ CFGWRT*LV WKQ HH+H R T T S RKNICSR V WD+
Sbjct: 416 K*DSFDSR*C*RYGATTCFGWRT*LVWPWKQGHHYHSRQTFTDQS*YRKNICSRGVVWDK 595
Query: 348 SS*IVEVEGF* 358
SS*IVE +GF*
Sbjct: 596 SS*IVEXDGF* 628
Score = 21.9 bits (45), Expect(2) = 4e-62
Identities = 7/7 (100%), Positives = 7/7 (100%)
Frame = +3
Query: 160 GYHDSPP 166
GYHDSPP
Sbjct: 3 GYHDSPP 23
>BI265594 weakly similar to GP|9965103|gb| resistance protein LM6 {Glycine
max}, partial (15%)
Length = 664
Score = 215 bits (548), Expect(2) = 1e-60
Identities = 111/145 (76%), Positives = 120/145 (82%)
Frame = +2
Query: 604 KKFQDMTILILDHCEYLTHIPDVSGLSNLEKLSFEYCKNLITIHNSIGHLNKLERLSAFG 663
+ FQ+M +L LD EYLTHIPD+SGL NLEK SF+YC+NLITI NSIGHLNKLERLSAFG
Sbjct: 179 QNFQNMKVLTLDDNEYLTHIPDLSGLQNLEKFSFKYCENLITIDNSIGHLNKLERLSAFG 358
Query: 664 CRTLKRFPPLGLASLKELKLSCCYSLKSFPKLLCKMTNIDKIWFWYTSIRELPSSFQNLS 723
C L+RFPPLGLASLKEL L CC SLKSFPKLLC+MTNID IW YT I EL SSFQNLS
Sbjct: 359 CSKLERFPPLGLASLKELNLCCCDSLKSFPKLLCEMTNIDCIWLNYTPIGELLSSFQNLS 538
Query: 724 ELDELSVREFGMLRFPKHNDRMYSI 748
ELDELSVRE GML ND+MYSI
Sbjct: 539 ELDELSVRECGML-----NDKMYSI 598
Score = 37.4 bits (85), Expect(2) = 1e-60
Identities = 21/26 (80%), Positives = 22/26 (83%)
Frame = +3
Query: 570 LSKSLSSSILSKASEIKSFSNCIYL* 595
LS+SLSSSILSKASEI SF IYL*
Sbjct: 3 LSESLSSSILSKASEITSFYKSIYL* 80
>BF636566 weakly similar to GP|9965103|gb| resistance protein LM6 {Glycine
max}, partial (20%)
Length = 663
Score = 229 bits (585), Expect = 2e-60
Identities = 134/212 (63%), Positives = 157/212 (73%)
Frame = +2
Query: 223 TCESNL*LYC*SI*MLMFS*KCEREFSFK*LEKSPAGAPFKNTST*N*VRKC*RRNS*NK 282
TC+SNL* C SI* MFS REF+ *LE SP GA FKN+ +*VR+C* NS K
Sbjct: 2 TCKSNL*FCCRSI*RCMFSS*S*REFNS**LETSPEGASFKNS*IEH*VRRC**GNSTYK 181
Query: 283 GKATWKEDSFDS**C*QTGSARCFGWRT*LVRSWKQSHHHHPR*TLTRLSWDRKNICSRR 342
GK EDSFDS *C*QTG+AR WRT*LV SWK SHH+H R*TLT + WDRKNIC +R
Sbjct: 182 GKTK*NEDSFDSR*C*QTGTARGISWRT*LVWSWK*SHHYHSR*TLTNMPWDRKNICGKR 361
Query: 343 VEWDRSS*IVEVEGF*K*ESSFKL*RHFKACSCICFWPSISDRSSGF*LVWKEYCRM*VY 402
DRS *IVEV+GF*K SSF+L*R FK CSCICF PS+S R+SGF*LVWKE+ RM ++
Sbjct: 362 AT*DRSF*IVEVDGF*KW*SSFEL*RCFKPCSCICFRPSVSSRNSGF*LVWKEHGRMAMH 541
Query: 403 IR*VWKNST*RYPKDT*TQL*CFGRRGAECLF 434
IR V K+S *R PKDT*++L*CF ++ ++ F
Sbjct: 542 IRWV*KDSQ*RDPKDT*SKL*CFWKKSSKVSF 637
Score = 29.3 bits (64), Expect = 5.8
Identities = 12/18 (66%), Positives = 13/18 (71%)
Frame = +3
Query: 425 FGRRGAECLFGYCLLHQR 442
FGRR A+CL YCL QR
Sbjct: 609 FGRRAAKCLSRYCLXFQR 662
>BF650588 weakly similar to GP|7341113|gb|A unknown {Glycine max}, partial
(64%)
Length = 645
Score = 202 bits (515), Expect(2) = 9e-59
Identities = 98/129 (75%), Positives = 110/129 (84%)
Frame = +3
Query: 37 KALTDKGINTFIDDNGLQRGNEITPSLLKAIEESRIFIPVFSINYASSSFCLDELDHIIH 96
+ L DKGI+TFIDD L+RG+EITPSL+KAIEESRIFIPVFSINYASS FCLDEL HIIH
Sbjct: 57 RLLLDKGIHTFIDDCDLKRGDEITPSLIKAIEESRIFIPVFSINYASSKFCLDELVHIIH 236
Query: 97 CYKTKGRPVLPVFFGVDPSHVRHHKGSYGEALAEHEKRFQNDPKNMERLQGWKDALSQAA 156
CYKTKGR VLPVF+GVDP+ +RH GSYGE L +HE+ FQN+ KN ERL WK AL+QAA
Sbjct: 237 CYKTKGRLVLPVFYGVDPTQIRHQSGSYGEHLTKHEESFQNNKKNKERLHQWKLALTQAA 416
Query: 157 NLSGYHDSP 165
NLSGYH SP
Sbjct: 417 NLSGYHYSP 443
Score = 43.5 bits (101), Expect(2) = 9e-59
Identities = 20/25 (80%), Positives = 23/25 (92%)
Frame = +2
Query: 19 FLSFRGSDTRYGFTGNLYKALTDKG 43
FL+FRGSDTR GFTG+LYKALT +G
Sbjct: 2 FLNFRGSDTRDGFTGHLYKALT*QG 76
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.347 0.151 0.545
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 29,677,394
Number of Sequences: 36976
Number of extensions: 503426
Number of successful extensions: 5866
Number of sequences better than 10.0: 218
Number of HSP's better than 10.0 without gapping: 2392
Number of HSP's successfully gapped in prelim test: 255
Number of HSP's that attempted gapping in prelim test: 3193
Number of HSP's gapped (non-prelim): 2885
length of query: 751
length of database: 9,014,727
effective HSP length: 103
effective length of query: 648
effective length of database: 5,206,199
effective search space: 3373616952
effective search space used: 3373616952
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 14 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.8 bits)
S2: 62 (28.5 bits)
Medicago: description of AC138133.8


