

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC138133.14 + phase: 0
(576 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
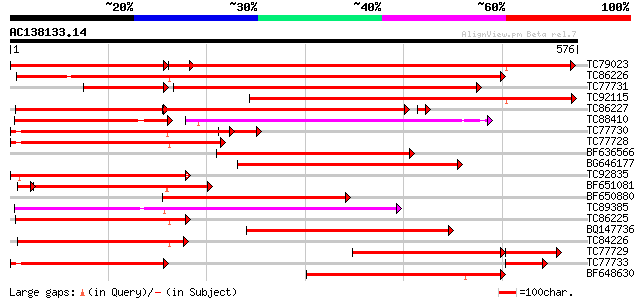
Score E
Sequences producing significant alignments: (bits) Value
TC79023 weakly similar to GP|18033111|gb|AAL56987.1 functional c... 822 0.0
TC86226 weakly similar to GP|18033111|gb|AAL56987.1 functional c... 710 0.0
TC77731 weakly similar to GP|18033111|gb|AAL56987.1 functional c... 467 e-132
TC92115 weakly similar to GP|18033111|gb|AAL56987.1 functional c... 435 e-122
TC86227 weakly similar to GP|18033111|gb|AAL56987.1 functional c... 351 3e-98
TC88410 weakly similar to GP|12056928|gb|AAG48132.1 putative res... 188 1e-87
TC77730 weakly similar to GP|7341113|gb|AAF61210.1| unknown {Gly... 311 4e-85
TC77728 similar to GP|12056928|gb|AAG48132.1 putative resistance... 298 3e-81
BF636566 weakly similar to GP|9965103|gb| resistance protein LM6... 295 2e-80
BG646177 weakly similar to GP|18033111|g functional candidate re... 281 4e-76
TC92835 weakly similar to GP|18033111|gb|AAL56987.1 functional c... 268 6e-72
BF651081 weakly similar to GP|18033111|g functional candidate re... 256 3e-71
BF650880 weakly similar to GP|9858476|gb| resistance protein LM1... 265 5e-71
TC89385 similar to GP|19774173|gb|AAL99063.1 NBS-LRR-Toll resist... 257 7e-69
TC86225 weakly similar to GP|9858476|gb|AAG01051.1| resistance p... 253 1e-67
BQ147736 weakly similar to GP|18033111|g functional candidate re... 247 1e-65
TC84226 similar to GP|22037313|gb|AAM89998.1 disease resistance-... 246 2e-65
TC77729 weakly similar to GP|22037342|gb|AAM90012.1 disease resi... 245 3e-65
TC77733 weakly similar to GP|18033111|gb|AAL56987.1 functional c... 237 8e-63
BF648630 weakly similar to GP|18033111|g functional candidate re... 228 4e-60
>TC79023 weakly similar to GP|18033111|gb|AAL56987.1 functional candidate
resistance protein KR1 {Glycine max}, partial (37%)
Length = 2072
Score = 822 bits (2123), Expect = 0.0
Identities = 413/421 (98%), Positives = 413/421 (98%), Gaps = 8/421 (1%)
Frame = +3
Query: 162 ISREPLDVAKYPVGLQSRVQHVKGHLDEKSDDEVHMVGLYGTGGIGKSTLAKAIYNFIAD 221
ISREPLDVAKYPVGLQSRVQHVKGHLDEKSDDEVHMVGLYGTGGIGKSTLAKAIYNFIAD
Sbjct: 738 ISREPLDVAKYPVGLQSRVQHVKGHLDEKSDDEVHMVGLYGTGGIGKSTLAKAIYNFIAD 917
Query: 222 QFEVLCFLENVRVNSTSDNLKHLQEKLLLKTVRLDIKLGGVSQGIPIIKQRLCRKKILLI 281
QFEVLCFLENVRVNSTSDNLKHLQEKLLLKTVRLDIKLGGVSQGIPIIKQRLCRKKILLI
Sbjct: 918 QFEVLCFLENVRVNSTSDNLKHLQEKLLLKTVRLDIKLGGVSQGIPIIKQRLCRKKILLI 1097
Query: 282 LDDVDKLDQLEALAGGLDWFGPGSRVIITTRNKHLLKIHGIESTHAVEGLNATEALELLR 341
LDDVDKLDQLEALAGGLDWFGPGSRVIITTRNKHLLKIHGIESTHAVEGLNATEALELLR
Sbjct: 1098 LDDVDKLDQLEALAGGLDWFGPGSRVIITTRNKHLLKIHGIESTHAVEGLNATEALELLR 1277
Query: 342 WMAFKENVPSSHEDILNRALTYASGLPLAIVIIGSNLVGRSVQDSMSTLDGYEEIPNKEI 401
WMAFKENVPSSHEDILNRALTYASGLPLAIVIIGSNLVGRSVQDSMSTLDGYEEIPNKEI
Sbjct: 1278 WMAFKENVPSSHEDILNRALTYASGLPLAIVIIGSNLVGRSVQDSMSTLDGYEEIPNKEI 1457
Query: 402 QRILKVSYDSLEKEEQSVFLDIACCFKGCKWPEVKEILHAHYGHCIVHHVAVLAEKSLMD 461
QRILKVSYDSLEKEEQSVFLDIACCFKGCKWPEVKEILHAHYGHCIVHHVAVLAEKSLMD
Sbjct: 1458 QRILKVSYDSLEKEEQSVFLDIACCFKGCKWPEVKEILHAHYGHCIVHHVAVLAEKSLMD 1637
Query: 462 HLKYDSYVTLHDLIEDMGKEVVRQESPDEPGERSRLWFERDI--------GTRKIKMINM 513
HLKYDSYVTLHDLIEDMGKEVVRQESPDEPGERSRLWFERDI GTRKIKMINM
Sbjct: 1638 HLKYDSYVTLHDLIEDMGKEVVRQESPDEPGERSRLWFERDIVHVLKKNTGTRKIKMINM 1817
Query: 514 KFPSMESDIDWNGNAFEKMTNLKTFITENGHHSKSLEYLPSSLRVMKGCIPKSPSSSSSN 573
KFPSMESDIDWNGNAFEKMTNLKTFITENGHHSKSLEYLPSSLRVMKGCIPKSPSSSSSN
Sbjct: 1818 KFPSMESDIDWNGNAFEKMTNLKTFITENGHHSKSLEYLPSSLRVMKGCIPKSPSSSSSN 1997
Query: 574 K 574
K
Sbjct: 1998 K 2000
Score = 335 bits (860), Expect(2) = 7e-97
Identities = 161/161 (100%), Positives = 161/161 (100%)
Frame = +1
Query: 1 MATQSPSSFTYQVFLSFRGADTRHGFTGNLYKALTDKGIYTFIDDNDLQRGDEITPSLKN 60
MATQSPSSFTYQVFLSFRGADTRHGFTGNLYKALTDKGIYTFIDDNDLQRGDEITPSLKN
Sbjct: 64 MATQSPSSFTYQVFLSFRGADTRHGFTGNLYKALTDKGIYTFIDDNDLQRGDEITPSLKN 243
Query: 61 AIEKSRIFIPVFSENYASSSFCLDELVHITHCYDTKGCLVLPVFIGVDPTDVRHHTGRYG 120
AIEKSRIFIPVFSENYASSSFCLDELVHITHCYDTKGCLVLPVFIGVDPTDVRHHTGRYG
Sbjct: 244 AIEKSRIFIPVFSENYASSSFCLDELVHITHCYDTKGCLVLPVFIGVDPTDVRHHTGRYG 423
Query: 121 EALAVHKKKFQNDKDNTERLQQWKEALSQAANLSGQHYKHG 161
EALAVHKKKFQNDKDNTERLQQWKEALSQAANLSGQHYKHG
Sbjct: 424 EALAVHKKKFQNDKDNTERLQQWKEALSQAANLSGQHYKHG 546
Score = 37.4 bits (85), Expect(2) = 7e-97
Identities = 17/26 (65%), Positives = 21/26 (80%)
Frame = +2
Query: 162 ISREPLDVAKYPVGLQSRVQHVKGHL 187
I+R+P DVAKY V LQ +VQH+K HL
Sbjct: 596 INRKPQDVAKYHVELQLQVQHIKTHL 673
>TC86226 weakly similar to GP|18033111|gb|AAL56987.1 functional candidate
resistance protein KR1 {Glycine max}, partial (24%)
Length = 1769
Score = 710 bits (1832), Expect = 0.0
Identities = 363/516 (70%), Positives = 415/516 (80%), Gaps = 20/516 (3%)
Frame = +3
Query: 8 SFTYQVFLSFRGADTRHGFTGNLYKALTDKGIYTFIDDND--LQRGDEITPSLKNAIEKS 65
+F+Y VFL +G DTR+GFTGNL KAL DKGI TF DD+D LQR D++TP + IE+S
Sbjct: 81 AFSYDVFLICKGTDTRYGFTGNLLKALIDKGIRTFHDDDDSDLQRRDKVTPII---IEES 251
Query: 66 RIFIPVFSENYASSSFCLDELVHITHCYDTKGCLVLPVFIGVDPTDVRHHTGRYGEALAV 125
RI IP+FS NYASSS CLD LVHI HCY TKGCLVLPVF GV+PTDVRHHTGRYG+ALA
Sbjct: 252 RILIPIFSANYASSSSCLDTLVHIIHCYKTKGCLVLPVFFGVEPTDVRHHTGRYGKALAE 431
Query: 126 HKKKFQNDKDNTERLQQWKEALSQAANLSGQHY-KHG----------------ISREPLD 168
H+ +FQND N ERLQQWK ALS AANL H HG ISR+ L
Sbjct: 432 HENRFQNDTKNMERLQQWKVALSLAANLPSYHDDSHGYEYELIGKIVKYISNKISRQSLH 611
Query: 169 VAKYPVGLQSRVQHVKGHLDEKSDDEVHMVGLYGTGGIGKSTLAKAIYNFIADQFEVLCF 228
VA YPVGLQSRVQ VK LDE DD VHMVG+YG GG GKSTLA+AIYNF+ADQFE LCF
Sbjct: 612 VATYPVGLQSRVQQVKSLLDEGPDDGVHMVGIYGIGGSGKSTLARAIYNFVADQFEGLCF 791
Query: 229 LENVRVNSTSDNLKHLQEKLLLKTVRLDIKLGGVSQGIPIIKQRLCRKKILLILDDVDKL 288
LE VR NS S++LK QE LL KT++L IKL VS+GI IIK+RLCRKKILLILDDVD +
Sbjct: 792 LEQVRENSASNSLKRFQEMLLSKTLQLKIKLADVSEGISIIKERLCRKKILLILDDVDNM 971
Query: 289 DQLEALAGGLDWFGPGSRVIITTRNKHLLKIHGIESTHAVEGLNATEALELLRWMAFK-E 347
QL ALAGG+DWFGPGSRVIITTR+KHLL H IE T+AV+GLN TEALELLRWMAFK +
Sbjct: 972 KQLNALAGGVDWFGPGSRVIITTRDKHLLACHEIEKTYAVKGLNVTEALELLRWMAFKND 1151
Query: 348 NVPSSHEDILNRALTYASGLPLAIVIIGSNLVGRSVQDSMSTLDGYEEIPNKEIQRILKV 407
VPSS+E ILNR + YASGLP+ I I+GSNL G+++++ +TLD YE+IPNKEIQRILKV
Sbjct: 1152KVPSSYEKILNRVVAYASGLPVVIEIVGSNLFGKNIEECKNTLDWYEKIPNKEIQRILKV 1331
Query: 408 SYDSLEKEEQSVFLDIACCFKGCKWPEVKEILHAHYGHCIVHHVAVLAEKSLMDHLKYDS 467
SYDSLE+EEQSVFLDIACCFKGCKW +VKEILHAHYGHCI HHV VL EK L+DH +YDS
Sbjct: 1332SYDSLEEEEQSVFLDIACCFKGCKWEKVKEILHAHYGHCINHHVEVLVEKCLIDHFEYDS 1511
Query: 468 YVTLHDLIEDMGKEVVRQESPDEPGERSRLWFERDI 503
+V+LH+LIE+MGKE+VR ESP EPG+RSRLWFE+DI
Sbjct: 1512HVSLHNLIENMGKELVRLESPFEPGKRSRLWFEKDI 1619
>TC77731 weakly similar to GP|18033111|gb|AAL56987.1 functional candidate
resistance protein KR1 {Glycine max}, partial (23%)
Length = 1651
Score = 467 bits (1202), Expect = e-132
Identities = 237/314 (75%), Positives = 269/314 (85%), Gaps = 1/314 (0%)
Frame = +1
Query: 167 LDVAKYPVGLQSRVQHVKGHLDEKSDDEVHMVGLYGTGGIGKSTLAKAIYNFIADQFEVL 226
L+VAKYPVGLQSR++ VK LD S+DEV MVGL+GTGG+GKSTLAKA+YNF+ADQFE +
Sbjct: 562 LNVAKYPVGLQSRIEQVKLLLDMGSEDEVRMVGLFGTGGMGKSTLAKAVYNFVADQFEGV 741
Query: 227 CFLENVRVNSTSDNLKHLQEKLLLKTVRLDIKLGGVSQGIPIIKQRLCRKKILLILDDVD 286
CFL NVR NST NLKHLQ+KLL K V+ D KL VS+GIPIIK+RL RKKILLILDDVD
Sbjct: 742 CFLHNVRENSTHKNLKHLQKKLLSKIVKFDGKLEDVSEGIPIIKERLSRKKILLILDDVD 921
Query: 287 KLDQLEALAGGLDWFGPGSRVIITTRNKHLLKIHGIESTHAVEGLNATEALELLRWMAFK 346
KL+QLEALAGGLDWFG GSRVIITTR+KHLL HGI STHAVE LN TEALELLR MAFK
Sbjct: 922 KLEQLEALAGGLDWFGHGSRVIITTRDKHLLACHGITSTHAVEELNETEALELLRRMAFK 1101
Query: 347 -ENVPSSHEDILNRALTYASGLPLAIVIIGSNLVGRSVQDSMSTLDGYEEIPNKEIQRIL 405
+ VPS++E+ILNR +TYASGLPLAIV IG NL GR V+D LD YE IPNK+IQRIL
Sbjct: 1102NDKVPSTYEEILNRVVTYASGLPLAIVTIGDNLFGRKVEDWKRILDEYENIPNKDIQRIL 1281
Query: 406 KVSYDSLEKEEQSVFLDIACCFKGCKWPEVKEILHAHYGHCIVHHVAVLAEKSLMDHLKY 465
+VSYD+LE +E+SVFLDIACCFKGCKW +VK+ILHAHYGHCI HHV VLAEKSL+ H +Y
Sbjct: 1282QVSYDALEPKEKSVFLDIACCFKGCKWTKVKKILHAHYGHCIEHHVGVLAEKSLIGHWEY 1461
Query: 466 DSYVTLHDLIEDMG 479
D+ +TLHDLIEDMG
Sbjct: 1462DTQMTLHDLIEDMG 1503
Score = 129 bits (324), Expect = 3e-30
Identities = 60/87 (68%), Positives = 70/87 (79%), Gaps = 1/87 (1%)
Frame = +2
Query: 76 YASSSFCLDELVHITHCYDTKGCLVLPVFIGVDPTDVRHHTGRYGEALAVHKKKFQNDKD 135
YASSSFCLDELVHI HCY TK CLVLPVF V+PT +RH +G YGE L H+++FQN++
Sbjct: 2 YASSSFCLDELVHIIHCYKTKSCLVLPVFYDVEPTHIRHQSGSYGEYLTKHEERFQNNEK 181
Query: 136 NTERLQQWKEALSQAANLSGQHYK-HG 161
N ERL+QWK AL+QAANLSG HY HG
Sbjct: 182 NMERLRQWKIALTQAANLSGYHYSPHG 262
>TC92115 weakly similar to GP|18033111|gb|AAL56987.1 functional candidate
resistance protein KR1 {Glycine max}, partial (24%)
Length = 1348
Score = 435 bits (1119), Expect = e-122
Identities = 233/347 (67%), Positives = 272/347 (78%), Gaps = 15/347 (4%)
Frame = +2
Query: 244 LQEKLLLKTVRLDIKLGGVSQGIPIIKQRLCRKKILLILDDVDKLDQLEALAGGLDWFGP 303
LQE+LLLK ++L IKLGGVS+GIP IK+RL KK LLILDDVD ++QL ALAGG DWFG
Sbjct: 2 LQEELLLKALQLKIKLGGVSEGIPYIKERLHTKKTLLILDDVDDMEQLHALAGGPDWFGR 181
Query: 304 GSRVIITTRNKHLLKIHGIESTHAVEGLNATEALELLRWMAFKEN-VPSSHEDILNRALT 362
GSRVIITTR+KHLL+ HGIESTH VE L TEALELLRWMAFK N VPS +ED+LNRA++
Sbjct: 182 GSRVIITTRDKHLLRSHGIESTHEVEELYGTEALELLRWMAFKNNKVPSIYEDVLNRAVS 361
Query: 363 YASGLPLAIVIIGSNLVGRSVQDSMSTLDGYEEIPNKEIQRILKVSYDSLEKEEQSVFLD 422
YASGLPL + I+GSNL G+++++ TLDGYE+IPNK+I +ILKVSYD+LE+E+QSVFLD
Sbjct: 362 YASGLPLVLEIVGSNLFGKTIEEWKGTLDGYEKIPNKKIHQILKVSYDALEEEQQSVFLD 541
Query: 423 IACCFKGCKWPEVKEILHAHYGHCIVHHVAVLAEKSL--MDHLKYDSY--VTLHDLIEDM 478
IACCFKGC W E + IL AHYGH I HH+ VLAEKSL + H Y S +TLHDLI++M
Sbjct: 542 IACCFKGCGWEEFEYILRAHYGHRITHHLVVLAEKSLVKITHPHYGSIYELTLHDLIKEM 721
Query: 479 GKEVVRQESPDEPGERSRLWFERDI--------GTRKIKMINMKFPSMESDIDWNGNAFE 530
GKEVVRQESP EPGERSRLW E DI GT KI+MI M FPS E ID G AF+
Sbjct: 722 GKEVVRQESPKEPGERSRLWCEDDIVNVLKENTGTSKIEMIYMNFPSEEFVIDKKGKAFK 901
Query: 531 KMTNLKTFITENGHHSKSLEYLPSSLRVMK--GCIPKSPSSSSSNKA 575
KMT LKT I EN H SK L+YLPSSLRV+K GC+ +S S S +KA
Sbjct: 902 KMTRLKTLIIENVHFSKGLKYLPSSLRVLKLRGCLSESLISCSLSKA 1042
>TC86227 weakly similar to GP|18033111|gb|AAL56987.1 functional candidate
resistance protein KR1 {Glycine max}, partial (22%)
Length = 1736
Score = 351 bits (900), Expect(2) = 3e-98
Identities = 180/252 (71%), Positives = 211/252 (83%), Gaps = 1/252 (0%)
Frame = +1
Query: 156 QHYKHGISREPLDVAKYPVGLQSRVQHVKGHLDEKSDDEVHMVGLYGTGGIGKSTLAKAI 215
+H I+R L VAKYPVGLQSRVQ VK LDE+SD+ V+MVGLYGT +GKSTLAKAI
Sbjct: 712 EHISDRINRVFLHVAKYPVGLQSRVQQVKLLLDEESDEGVNMVGLYGTRALGKSTLAKAI 891
Query: 216 YNFIADQFEVLCFLENVRVNSTSDNLKHLQEKLLLKTVRLDIKLGGVSQGIPIIKQRLCR 275
YNFIADQFE +CFL NVR NS NLKHLQ++LL KTV+L+IKL VS+GIPIIK+RLCR
Sbjct: 892 YNFIADQFEGVCFLHNVRENSARKNLKHLQKELLSKTVQLNIKLRDVSEGIPIIKERLCR 1071
Query: 276 KKILLILDDVDKLDQLEALAGGLDWFGPGSRVIITTRNKHLLKIHGIESTHAVEGLNATE 335
KKILLILDDVD+LDQLEALAGGLDWFGPGSRVIITTR+KHLL HGIE T+AV GL E
Sbjct: 1072KKILLILDDVDQLDQLEALAGGLDWFGPGSRVIITTRDKHLLTCHGIERTYAVRGLYGKE 1251
Query: 336 ALELLRWMAFKEN-VPSSHEDILNRALTYASGLPLAIVIIGSNLVGRSVQDSMSTLDGYE 394
ALELLRW AFK N VP S+ED+LNRA++Y SG+PL + I+GSNL G++++ +TLDGY+
Sbjct: 1252ALELLRWTAFKNNKVPPSYEDVLNRAVSYGSGIPLVLEIVGSNLFGKNIEVWKNTLDGYD 1431
Query: 395 EIPNKEIQRILK 406
IPNKEIQ+IL+
Sbjct: 1432RIPNKEIQKILR 1467
Score = 235 bits (599), Expect = 4e-62
Identities = 112/155 (72%), Positives = 124/155 (79%)
Frame = +3
Query: 7 SSFTYQVFLSFRGADTRHGFTGNLYKALTDKGIYTFIDDNDLQRGDEITPSLKNAIEKSR 66
S F Y VFLSFR DT +GFTGNLYKAL DKGI TFIDDNDL+RGDE TPSL AIE+SR
Sbjct: 6 SGFKYHVFLSFRDIDTLYGFTGNLYKALIDKGIKTFIDDNDLERGDESTPSLVKAIEESR 185
Query: 67 IFIPVFSENYASSSFCLDELVHITHCYDTKGCLVLPVFIGVDPTDVRHHTGRYGEALAVH 126
I IP+FS NYASSSFCLDELVHI HCY T+GC VLPVF G DPT VRH TG YGE L H
Sbjct: 186 ILIPIFSANYASSSFCLDELVHIIHCYKTRGCSVLPVFYGADPTHVRHQTGSYGEHLTKH 365
Query: 127 KKKFQNDKDNTERLQQWKEALSQAANLSGQHYKHG 161
+ KFQN+K+N ERL++WK AL+QAAN SG H+ G
Sbjct: 366 EDKFQNNKENMERLKKWKMALTQAANFSGHHFSQG 470
Score = 26.6 bits (57), Expect(2) = 3e-98
Identities = 11/13 (84%), Positives = 12/13 (91%)
Frame = +2
Query: 415 EEQSVFLDIACCF 427
EEQ+VFLDIAC F
Sbjct: 1466 EEQNVFLDIACFF 1504
>TC88410 weakly similar to GP|12056928|gb|AAG48132.1 putative resistance
protein {Glycine max}, partial (8%)
Length = 2252
Score = 188 bits (477), Expect(2) = 1e-87
Identities = 121/319 (37%), Positives = 182/319 (56%), Gaps = 7/319 (2%)
Frame = +1
Query: 179 RVQHVKGHLDEK---SDDEVHMVGLYGTGGIGKSTLAKAIYNFIAD-QFEVLCFLENVRV 234
RV+ V +L+ DD V +VG+ G GIGK+TLA+ +Y+F +F+ CF +NV
Sbjct: 604 RVEKVMRYLNSSPRSDDDGVCLVGICGVPGIGKTTLARGVYHFGGGTEFDSCCFFDNVGE 783
Query: 235 NSTSDNLKHLQEKLLLKTV--RLDIKLGGVSQGIPIIKQRLCRKKILLILDDVDKLDQLE 292
L HLQ+ LL V V + + IK L +KK+ L+L+DV + L+
Sbjct: 784 YVKKHGLVHLQQMLLSAIVGHNNSTMFENVDERVWKIKHMLNQKKVFLVLEDVHDSEVLK 963
Query: 293 ALAGGLDWFGPGSRVIITTRNKHLLKIHGIESTHAVEGLNATEALELLRWMAFKE-NVPS 351
A+ +FG GS+VIIT R K L+ HGI+ + VE +N TEA +LL AF N+
Sbjct: 964 AIVKLSTFFGSGSKVIITAREKCFLEFHGIKRIYEVERMNKTEAFQLLNLKAFDSMNISP 1143
Query: 352 SHEDILNRALTYASGLPLAIVIIGSNLVGRSVQDSMSTLDGYEEIPNKEIQRILKVSYDS 411
H IL TYASG P + +IGS L G+S+++ S L Y+EI N++I++IL+VS+D+
Sbjct: 1144CHVTILEGLETYASGHPFILEMIGSYLSGKSMEECESALHQYKEISNRDIKKILQVSFDA 1323
Query: 412 LEKEEQSVFLDIACCFKGCKWPEVKEILHAHYGHCIVHHVAVLAEKSLMDHLKYDSYVTL 471
LEK +Q++ + IA + + V+ +LH YG C + + VL KSL+ + + +V +
Sbjct: 1324LEKSQQNMLIHIALHLREQELEMVENLLHRKYGVCPKYDIRVLLNKSLI-KINENGHVIV 1500
Query: 472 HDLIEDMGKEVVRQESPDE 490
H L +DM VR + P E
Sbjct: 1501HVLTQDM----VRDDIPVE 1545
Score = 154 bits (389), Expect(2) = 1e-87
Identities = 82/161 (50%), Positives = 105/161 (64%), Gaps = 1/161 (0%)
Frame = +3
Query: 6 PSSFTYQVFLSFRGADTRHGFTGNLYKALTDKGIYTFIDDNDLQRGDEITPSLKNAIEKS 65
P +F Y F+SFRG+DTR GF+G L K L D+G TF DD +L+RG +IT + AIE+S
Sbjct: 147 PKAFRYSTFISFRGSDTRCGFSGFLNKYLIDRGFRTFFDDGELERGTQITVEIPKAIEES 326
Query: 66 RIFIPVFSENYASSSFCLDELVHITHCYDT-KGCLVLPVFIGVDPTDVRHHTGRYGEALA 124
RIFIPV SENYASSSFCLDELV I + G V PVF ++ +DV++ TG YG+ALA
Sbjct: 327 RIFIPVLSENYASSSFCLDELVKILEEFKKGNGRWVFPVFYYINISDVKNQTGSYGQALA 506
Query: 125 VHKKKFQNDKDNTERLQQWKEALSQAANLSGQHYKHGISRE 165
VHK + ER ++W AL+ A+ G H + SRE
Sbjct: 507 VHKNRVM-----PERFEKWINALASVADFRGCHMERSSSRE 614
>TC77730 weakly similar to GP|7341113|gb|AAF61210.1| unknown {Glycine max},
partial (80%)
Length = 831
Score = 311 bits (797), Expect = 4e-85
Identities = 161/245 (65%), Positives = 187/245 (75%), Gaps = 17/245 (6%)
Frame = +1
Query: 1 MATQSPSSFTYQVFLSFRGADTRHGFTGNLYKALTDKGIYTFIDDNDLQRGDEITPSLKN 60
MA QSPS +VFL+FRG+DTR+ FTGNLYKAL DKGI TFID+NDLQRGDEITPSL
Sbjct: 16 MAMQSPS----RVFLNFRGSDTRNNFTGNLYKALVDKGIRTFIDENDLQRGDEITPSLVK 183
Query: 61 AIEKSRIFIPVFSENYASSSFCLDELVHITHCYDTKGCLVLPVFIGVDPTDVRHHTGRYG 120
AIE+SRI IP+FS NYASSSFCLDELVHI HCY TK CLVLPVF GVDPTDVR HT YG
Sbjct: 184 AIEESRICIPIFSANYASSSFCLDELVHIIHCYKTKSCLVLPVFYGVDPTDVRRHTCSYG 363
Query: 121 EALAVHKKKFQNDKDNTERLQQWKEALSQAANLSGQHY-----------------KHGIS 163
E L H++ FQN++ N ERL+QWK AL+QAANLSG HY + I+
Sbjct: 364 EYLTKHEEGFQNNEKNMERLRQWKMALTQAANLSGYHYSPHEYEHKFIEKIVQYISNNIN 543
Query: 164 REPLDVAKYPVGLQSRVQHVKGHLDEKSDDEVHMVGLYGTGGIGKSTLAKAIYNFIADQF 223
+ L+VAKYPVGLQSR++ VK LD S+DEV MVGL+GTGG+GKSTLAKA++NF+AD
Sbjct: 544 HDFLNVAKYPVGLQSRIEQVKLLLDMGSEDEVRMVGLFGTGGMGKSTLAKAVFNFLADHL 723
Query: 224 EVLCF 228
+V F
Sbjct: 724 KVYVF 738
Score = 42.7 bits (99), Expect = 4e-04
Identities = 22/43 (51%), Positives = 29/43 (67%)
Frame = +3
Query: 213 KAIYNFIADQFEVLCFLENVRVNSTSDNLKHLQEKLLLKTVRL 255
K+ + F FE +CFL NVR NS+ +NLKHLQ+ LL K + L
Sbjct: 690 KSSF*FSC*SFEGVCFLHNVRENSSHNNLKHLQKMLLSK*LNL 818
>TC77728 similar to GP|12056928|gb|AAG48132.1 putative resistance protein
{Glycine max}, partial (12%)
Length = 765
Score = 298 bits (764), Expect = 3e-81
Identities = 156/236 (66%), Positives = 181/236 (76%), Gaps = 17/236 (7%)
Frame = +1
Query: 1 MATQSPSSFTYQVFLSFRGADTRHGFTGNLYKALTDKGIYTFIDDNDLQRGDEITPSLKN 60
MA QSPS +VFLSFRG+DTR+ FTGNLYKAL DKGI TFIDDNDL+RGDEITPSL
Sbjct: 70 MAMQSPS----RVFLSFRGSDTRNKFTGNLYKALVDKGIRTFIDDNDLERGDEITPSLVK 237
Query: 61 AIEKSRIFIPVFSENYASSSFCLDELVHITHCYDTKGCLVLPVFIGVDPTDVRHHTGRYG 120
AIE+SRIFIP+FS NYASSSFCLDELVHI HCY TK CLV PVF V+PT +R+ +G YG
Sbjct: 238 AIEESRIFIPIFSANYASSSFCLDELVHIIHCYKTKSCLVFPVFYDVEPTHIRNQSGIYG 417
Query: 121 EALAVHKKKFQNDKDNTERLQQWKEALSQAANLSGQHYK-HG----------------IS 163
E L H+++FQN++ N ERL+QWK AL QAANLSG HY HG I+
Sbjct: 418 EHLTKHEERFQNNEKNMERLRQWKIALIQAANLSGYHYSPHGYEYKFIEKIVEDISNNIN 597
Query: 164 REPLDVAKYPVGLQSRVQHVKGHLDEKSDDEVHMVGLYGTGGIGKSTLAKAIYNFI 219
L+VAKYPVGLQSR++ VK LD S+DEV MVGL+GTGG+GKSTLAKA+YNF+
Sbjct: 598 HVFLNVAKYPVGLQSRIEEVKLLLDMGSEDEVRMVGLFGTGGMGKSTLAKAVYNFV 765
>BF636566 weakly similar to GP|9965103|gb| resistance protein LM6 {Glycine
max}, partial (20%)
Length = 663
Score = 295 bits (756), Expect = 2e-80
Identities = 149/202 (73%), Positives = 175/202 (85%), Gaps = 1/202 (0%)
Frame = +3
Query: 211 LAKAIYNFIADQFEVLCFLENVRVNSTSDNLKHLQEKLLLKTVRLDIKLGGVSQGIPIIK 270
LAKAIYNF+ADQFE +CFL VR NST ++LKHLQ++LLLKTV+L+IKLG S+GIP+IK
Sbjct: 3 LAKAIYNFVADQFEGVCFLHKVRENSTHNSLKHLQKELLLKTVKLNIKLGDASEGIPLIK 182
Query: 271 QRLCRKKILLILDDVDKLDQLEALAGGLDWFGPGSRVIITTRNKHLLKIHGIESTHAVEG 330
+RL R KILLILDDVDKL+QLEALAGGLDWFG GSRVIITTR+KHLL HGIE T+AV G
Sbjct: 183 ERLNRMKILLILDDVDKLEQLEALAGGLDWFGHGSRVIITTRDKHLLTCHGIERTYAVNG 362
Query: 331 LNATEALELLRWMAFKE-NVPSSHEDILNRALTYASGLPLAIVIIGSNLVGRSVQDSMST 389
L+ TEA ELLRWMAFK VPSS+ D+LNRA+ YASGLPL + I+GSNL G+S+++ T
Sbjct: 363 LHETEAFELLRWMAFKNGEVPSSYNDVLNRAVAYASGLPLVLEIVGSNLFGKSMEEWQCT 542
Query: 390 LDGYEEIPNKEIQRILKVSYDS 411
LDGYE+IPNKEIQRILKVSYD+
Sbjct: 543 LDGYEKIPNKEIQRILKVSYDA 608
Score = 38.9 bits (89), Expect = 0.005
Identities = 20/38 (52%), Positives = 25/38 (65%)
Frame = +1
Query: 392 GYEEIPNKEIQRILKVSYDSLEKEEQSVFLDIACCFKG 429
G + P K + LK + LE+E+QSVFLDIA CFKG
Sbjct: 550 GMKRFPIKRSKGYLK*AMMLLEEEQQSVFLDIAXCFKG 663
>BG646177 weakly similar to GP|18033111|g functional candidate resistance
protein KR1 {Glycine max}, partial (17%)
Length = 708
Score = 281 bits (720), Expect = 4e-76
Identities = 144/230 (62%), Positives = 180/230 (77%), Gaps = 1/230 (0%)
Frame = +3
Query: 232 VRVNSTSDNLKHLQEKLLLKTVRLDIKLGGVSQGIPIIKQRLCRKKILLILDDVDKLDQL 291
VR S L+ LQEKLL KTV L +K G VS+GIPIIK+RL KK+LLILDDVD+L QL
Sbjct: 3 VREISAKHGLEDLQEKLLSKTVGLSVKFGHVSEGIPIIKERLRLKKVLLILDDVDELKQL 182
Query: 292 EALAGGLDWFGPGSRVIITTRNKHLLKIHGIESTHAVEGLNATEALELLRWMAFKEN-VP 350
+ LAG +W G GSRV++TTR+KHLL HGIE T+ ++GLN EALELL+W AFK N V
Sbjct: 183 KVLAGDPNWLGHGSRVVVTTRDKHLLACHGIERTYELDGLNKEEALELLKWKAFKNNKVD 362
Query: 351 SSHEDILNRALTYASGLPLAIVIIGSNLVGRSVQDSMSTLDGYEEIPNKEIQRILKVSYD 410
SS+E ILNRA+TYASGLPLA+ ++GS+L G+ + STLD YE IP+KE+ +ILKVS+D
Sbjct: 363 SSYEHILNRAVTYASGLPLALEVVGSSLFGKHKDEWKSTLDRYERIPHKEVLKILKVSFD 542
Query: 411 SLEKEEQSVFLDIACCFKGCKWPEVKEILHAHYGHCIVHHVAVLAEKSLM 460
SLEK+EQSVFLDIACCF+G EV++IL+AHYG C+ +H+ VL EK L+
Sbjct: 543 SLEKDEQSVFLDIACCFRGYILAEVEDILYAHYGECMKYHIRVLIEKCLI 692
>TC92835 weakly similar to GP|18033111|gb|AAL56987.1 functional candidate
resistance protein KR1 {Glycine max}, partial (13%)
Length = 581
Score = 268 bits (684), Expect = 6e-72
Identities = 141/191 (73%), Positives = 151/191 (78%), Gaps = 8/191 (4%)
Frame = +2
Query: 1 MATQSPSS-------FTYQVFLSFRGADTRHGFTGNLYKALTDKGIYTFIDDNDLQRGDE 53
MA Q PSS FTYQVFLSFRG DTRHGFTGNLYKALTDKGI TFIDDNDLQRGDE
Sbjct: 11 MAMQLPSSSSSFSSAFTYQVFLSFRGTDTRHGFTGNLYKALTDKGIKTFIDDNDLQRGDE 190
Query: 54 ITPSLKNAIEKSRIFIPVFSENYASSSFCLDELVHITHCYDTKGCLVLPVFIGVDPTDVR 113
ITPSL AIE+SRIFIPVFS NYA+S FCLDELVHI HCY TKG LVLPVF GVDPT+VR
Sbjct: 191 ITPSLLKAIEESRIFIPVFSINYATSKFCLDELVHIIHCYKTKGRLVLPVFFGVDPTNVR 370
Query: 114 HHTGRYGEALAVHKKKFQNDKDNTERLQQWKEALSQAANLSGQHYKHGISREPL-DVAKY 172
HHTG YGEALA H+K+FQNDKDN ERL QWK AL+QAANLSG H G + + D+ KY
Sbjct: 371 HHTGPYGEALAGHEKRFQNDKDNMERLHQWKLALTQAANLSGYHSSPGYEYKFIGDIVKY 550
Query: 173 PVGLQSRVQHV 183
SR QH+
Sbjct: 551 ISNKISR-QHL 580
>BF651081 weakly similar to GP|18033111|g functional candidate resistance
protein KR1 {Glycine max}, partial (16%)
Length = 664
Score = 256 bits (653), Expect(2) = 3e-71
Identities = 135/199 (67%), Positives = 150/199 (74%), Gaps = 17/199 (8%)
Frame = +1
Query: 25 GFTGNLYKALTDKGIYTFIDDNDLQRGDEITPSLKNAIEKSRIFIPVFSENYASSSFCLD 84
GFTGNLYKAL DKGI+TFID+++LQRGDEITPSL AIE+SRIFI VFS NYASSSFCLD
Sbjct: 67 GFTGNLYKALDDKGIHTFIDNHELQRGDEITPSLLKAIEESRIFIAVFSINYASSSFCLD 246
Query: 85 ELVHITHCYDTKGCLVLPVFIGVDPTDVRHHTGRYGEALAVHKKKFQNDKDNTERLQQWK 144
ELVHI HCY TKG LVLPVF V+PT VRH G YGEALA H+K+FQND + ERLQ WK
Sbjct: 247 ELVHIIHCYKTKGRLVLPVFFAVEPTIVRHQKGSYGEALAEHEKRFQNDPKSMERLQGWK 426
Query: 145 EALSQAANLSGQH-------YK----------HGISREPLDVAKYPVGLQSRVQHVKGHL 187
EALSQAANLSG H YK + IS++PL VA YPVGLQS Q +K L
Sbjct: 427 EALSQAANLSGYHDSPPGYEYKLIGKIVKYIXNKISQQPLHVATYPVGLQSXXQQMKSLL 606
Query: 188 DEKSDDEVHMVGLYGTGGI 206
DE SD VHMVG+YG GG+
Sbjct: 607 DEXSDHGVHMVGIYGIGGL 663
Score = 31.6 bits (70), Expect(2) = 3e-71
Identities = 12/18 (66%), Positives = 16/18 (88%)
Frame = +3
Query: 9 FTYQVFLSFRGADTRHGF 26
+ YQVFL+FRG+DTR+ F
Sbjct: 18 YKYQVFLNFRGSDTRYXF 71
>BF650880 weakly similar to GP|9858476|gb| resistance protein LM12 {Glycine
max}, partial (41%)
Length = 659
Score = 265 bits (676), Expect = 5e-71
Identities = 136/191 (71%), Positives = 154/191 (80%)
Frame = +3
Query: 156 QHYKHGISREPLDVAKYPVGLQSRVQHVKGHLDEKSDDEVHMVGLYGTGGIGKSTLAKAI 215
++ + I+R+PL VA YPVGL SRVQ VK LDE DD VHMVG+YG GG+GKS LA+AI
Sbjct: 57 KYISNKINRQPLHVANYPVGLHSRVQEVKSLLDEGPDDGVHMVGIYGIGGLGKSALARAI 236
Query: 216 YNFIADQFEVLCFLENVRVNSTSDNLKHLQEKLLLKTVRLDIKLGGVSQGIPIIKQRLCR 275
YNF+ADQFE LCFL +VR NS +NLKHLQEKLLLKT L IKL V +GIPIIK+RLCR
Sbjct: 237 YNFVADQFEGLCFLHDVRENSAQNNLKHLQEKLLLKTTGLKIKLDHVCEGIPIIKERLCR 416
Query: 276 KKILLILDDVDKLDQLEALAGGLDWFGPGSRVIITTRNKHLLKIHGIESTHAVEGLNATE 335
KILLILDDVD ++QL ALAGG DWFG GSRVIITTR+KHLL H IE T+AVEGL T+
Sbjct: 417 NKILLILDDVDDMEQLHALAGGPDWFGHGSRVIITTRDKHLLTSHDIERTYAVEGLYGTK 596
Query: 336 ALELLRWMAFK 346
ALELLR MAFK
Sbjct: 597 ALELLRXMAFK 629
>TC89385 similar to GP|19774173|gb|AAL99063.1 NBS-LRR-Toll resistance gene
analog protein {Medicago ruthenica}, partial (98%)
Length = 1285
Score = 257 bits (657), Expect = 7e-69
Identities = 149/413 (36%), Positives = 243/413 (58%), Gaps = 20/413 (4%)
Frame = +3
Query: 6 PSSFTYQVFLSFRGADTRHGFTGNLYKALTDKGIYTFIDDNDLQRGDEITPSLKNAIEKS 65
P Y VF++FRG DTR+ FT L+ AL KGIY F DD +L +G+ I P L IE S
Sbjct: 54 PRRNCYDVFVTFRGEDTRNNFTNFLFAALERKGIYAFRDDTNLPKGESIGPELLRTIEGS 233
Query: 66 RIFIPVFSENYASSSFCLDELVHITHCYDTKGCLVLPVFIGVDPTDVRHHTGRYGEALAV 125
++F+ V S NYASS++CL EL I C G VLP+F GVDP++V+ +G Y + A
Sbjct: 234 QVFVAVLSRNYASSTWCLQELEKICECIKGSGKYVLPIFYGVDPSEVKKQSGIYWDDFAK 413
Query: 126 HKKKFQNDKDNTERLQQWKEALSQAANLSG----------------QHYKHGISREPLDV 169
H+++F+ D ++ +W+EAL+Q +++G Q + + + V
Sbjct: 414 HEQRFKQD---PHKVSRWREALNQVGSIAGWDLRDKQQSVEVEKIVQTILNILKCKSSFV 584
Query: 170 AKYPVGLQSRVQHVKGHLDEKSDDEVHMVGLYGTGGIGKSTLAKAIYNFIADQFEVLCFL 229
+K VG+ SR + +K L S D V ++G++G GG GK+TLA +Y I +F+ CF+
Sbjct: 585 SKDLVGINSRTEALKHQLLLNSVDGVRVIGIWGMGGKGKTTLAMNLYGQICHRFDASCFI 764
Query: 230 ENV-RVNSTSDNLKHLQEKLLLKTVRLD-IKLGGVSQGIPIIKQRLCRKKILLILDDVDK 287
++V ++ D Q+++L +T+ ++ ++ +I+ RL R+K LLILD+VD+
Sbjct: 765 DDVSKIFRLHDGPIDAQKQILHQTLGIEHHQICNHYSATDLIRHRLSREKTLLILDNVDQ 944
Query: 288 LDQLEALAGGLDWFGPGSRVIITTRNKHLLKIHGIESTHAVEGLNATEALELLRWMAFK- 346
++QLE + +W G GSR++I +R++H+LK + ++ + V L+ TE+ +L R AFK
Sbjct: 945 VEQLERIGVHREWLGAGSRIVIISRDEHILKEYKVDVDYKVPLLDWTESHKLFRQKAFKL 1124
Query: 347 -ENVPSSHEDILNRALTYASGLPLAIVIIGSNLVGRSVQDSMSTLDGYEEIPN 398
+ + +++++ L YA+GLPL I ++GS L GR+V + S L + PN
Sbjct: 1125EKIIMKNYQNLAYEILNYANGLPLTITVLGSFLSGRNVTEWKSALARLRQRPN 1283
>TC86225 weakly similar to GP|9858476|gb|AAG01051.1| resistance protein LM12
{Glycine max}, partial (25%)
Length = 677
Score = 253 bits (647), Expect = 1e-67
Identities = 129/193 (66%), Positives = 142/193 (72%), Gaps = 16/193 (8%)
Frame = +3
Query: 7 SSFTYQVFLSFRGADTRHGFTGNLYKALTDKGIYTFIDDNDLQRGDEITPSLKNAIEKSR 66
S F Y VFLSFR DT +GFTGNLYKAL DKGI TFIDDNDL+RGDE TPSL AIE+SR
Sbjct: 99 SGFKYHVFLSFRDIDTLYGFTGNLYKALIDKGIKTFIDDNDLERGDESTPSLVKAIEESR 278
Query: 67 IFIPVFSENYASSSFCLDELVHITHCYDTKGCLVLPVFIGVDPTDVRHHTGRYGEALAVH 126
I IP+FS NYASSSFCLDELVHI HCY T+GC VLPVF G DPT VRH TG YGE L H
Sbjct: 279 ILIPIFSANYASSSFCLDELVHIIHCYKTRGCSVLPVFYGADPTHVRHQTGSYGEHLTKH 458
Query: 127 KKKFQNDKDNTERLQQWKEALSQAANLSGQHYKHG----------------ISREPLDVA 170
+ KFQN+K+N ERL++WK AL+QAAN SG H+ G I+R L VA
Sbjct: 459 EDKFQNNKENMERLKKWKMALTQAANFSGHHFSQGYEYELIENIVEHISDRINRVFLHVA 638
Query: 171 KYPVGLQSRVQHV 183
KYPVGLQSRVQ V
Sbjct: 639 KYPVGLQSRVQQV 677
>BQ147736 weakly similar to GP|18033111|g functional candidate resistance
protein KR1 {Glycine max}, partial (17%)
Length = 712
Score = 247 bits (630), Expect = 1e-65
Identities = 126/212 (59%), Positives = 162/212 (75%), Gaps = 1/212 (0%)
Frame = +2
Query: 241 LKHLQEKLLLKTVRLDIKLGGVSQGIPIIKQRLCRKKILLILDDVDKLDQLEALAGGLDW 300
L+HLQ KLL K V LDIKL V++GIPIIKQRL RKK+LLILDDV +L QL+ LAGGLDW
Sbjct: 50 LEHLQGKLLSKLVELDIKLVDVNEGIPIIKQRLHRKKVLLILDDVHELKQLQVLAGGLDW 229
Query: 301 FGPGSRVIITTRNKHLLKIHGIESTHAVEGLNATEALELLRWMAFKEN-VPSSHEDILNR 359
FGPGS+VIITTR++ LL HGIE + ++ LN EALELLRW AFK N V + +++L+
Sbjct: 230 FGPGSKVIITTRDEQLLAGHGIERAYEIDKLNEKEALELLRWNAFKNNKVNPNFDNVLHL 409
Query: 360 ALTYASGLPLAIVIIGSNLVGRSVQDSMSTLDGYEEIPNKEIQRILKVSYDSLEKEEQSV 419
A+TYASGLPLA+ ++GSNL G+++ + S L+ YE IP K+IQ ILKVS+D+L ++ +V
Sbjct: 410 AVTYASGLPLALEVVGSNLFGKNIGEWKSALNQYERIPIKKIQEILKVSFDALXEDXXNV 589
Query: 420 FLDIACCFKGCKWPEVKEILHAHYGHCIVHHV 451
FLDI+C K C V++ILHAHYG C H +
Sbjct: 590 FLDISCFLKXCDLKXVEDILHAHYGSCXKHQL 685
>TC84226 similar to GP|22037313|gb|AAM89998.1 disease resistance-like
protein GS0-1 {Glycine max}, partial (72%)
Length = 665
Score = 246 bits (627), Expect = 2e-65
Identities = 129/213 (60%), Positives = 147/213 (68%), Gaps = 40/213 (18%)
Frame = +3
Query: 9 FTYQVFLSFRGADTRHGFTGNLYKALTDKGIYTFIDDNDLQRGDEITPSLKNAIEKSRIF 68
F YQVFLSFRG+DTR+GFTGNLYKALTDKGI+TFIDD++LQRGDEITPSL NAIE+SRIF
Sbjct: 24 FKYQVFLSFRGSDTRYGFTGNLYKALTDKGIHTFIDDSELQRGDEITPSLDNAIEESRIF 203
Query: 69 IPVFSENYASSSFCLDELVHITHCYDTKGCLVLPVFIGVDPTDVRHHTGRYGEALAVHKK 128
IPVFS NYASSSFCLDELVHI H Y G LVLPVF GVDP+ VRHH G YGEALA H++
Sbjct: 204 IPVFSANYASSSFCLDELVHIIHLYKQNGRLVLPVFFGVDPSHVRHHRGSYGEALAKHEE 383
Query: 129 KFQNDKDNTERLQQWKEALSQAANLSGQHYKHG--------------------------- 161
+FQ++ D+ ERLQ+WK AL+QAANLSG H G
Sbjct: 384 RFQHNTDHMERLQKWKIALTQAANLSGDHRSPGYEYKLTGKIAFNQTPHLSSDCSQRYEY 563
Query: 162 -------------ISREPLDVAKYPVGLQSRVQ 181
I+R PL VA YPVG + R+Q
Sbjct: 564 DFIGDIVKYISNKINRVPLHVANYPVGFKFRIQ 662
>TC77729 weakly similar to GP|22037342|gb|AAM90012.1 disease resistance-like
protein GS3-5 {Glycine max}, partial (72%)
Length = 973
Score = 245 bits (626), Expect = 3e-65
Identities = 116/155 (74%), Positives = 133/155 (84%)
Frame = +1
Query: 349 VPSSHEDILNRALTYASGLPLAIVIIGSNLVGRSVQDSMSTLDGYEEIPNKEIQRILKVS 408
VPS++E+ILNR +TYASGLPLAIV IG NL GR V+D LD YE IPNK+IQRIL+VS
Sbjct: 1 VPSTYEEILNRVVTYASGLPLAIVTIGDNLFGRKVEDWKRILDEYENIPNKDIQRILQVS 180
Query: 409 YDSLEKEEQSVFLDIACCFKGCKWPEVKEILHAHYGHCIVHHVAVLAEKSLMDHLKYDSY 468
YD+LE +E+SVFLDIACCFKGCKW +VK+ILHAHYGHCI HHV VLAEKSL+ H +YD+
Sbjct: 181 YDALEPKEKSVFLDIACCFKGCKWTKVKKILHAHYGHCIEHHVGVLAEKSLIGHWEYDTQ 360
Query: 469 VTLHDLIEDMGKEVVRQESPDEPGERSRLWFERDI 503
+TLHDLIEDMGKE+VRQESP PGERSRLWF DI
Sbjct: 361 MTLHDLIEDMGKEIVRQESPKNPGERSRLWFHDDI 465
Score = 52.8 bits (125), Expect = 4e-07
Identities = 26/57 (45%), Positives = 36/57 (62%)
Frame = +2
Query: 504 GTRKIKMINMKFPSMESDIDWNGNAFEKMTNLKTFITENGHHSKSLEYLPSSLRVMK 560
GT I+MI +K+ + +W+G AF KMTNLKT I ++ S YLPSSLR ++
Sbjct: 653 GTENIEMIYLKYGLTARETEWDGMAFNKMTNLKTLIIDDYKFSGGPGYLPSSLRYLE 823
>TC77733 weakly similar to GP|18033111|gb|AAL56987.1 functional candidate
resistance protein KR1 {Glycine max}, partial (12%)
Length = 632
Score = 237 bits (605), Expect = 8e-63
Identities = 118/162 (72%), Positives = 133/162 (81%), Gaps = 1/162 (0%)
Frame = +3
Query: 1 MATQSPSSFTYQVFLSFRGADTRHGFTGNLYKALTDKGIYTFIDDNDLQRGDEITPSLKN 60
MA QSPS +VFLSFRG+DTR+ FTGNLYKAL DKGI TFIDDNDL+RGDEITPSL
Sbjct: 30 MAMQSPS----RVFLSFRGSDTRNNFTGNLYKALVDKGIRTFIDDNDLERGDEITPSLVK 197
Query: 61 AIEKSRIFIPVFSENYASSSFCLDELVHITHCYDTKGCLVLPVFIGVDPTDVRHHTGRYG 120
AIE+SRIFIP+FS NYASSSFCLDELVHI HCY TK CLVLPVF V+PT +RH +G YG
Sbjct: 198 AIEESRIFIPIFSANYASSSFCLDELVHIIHCYKTKSCLVLPVFYDVEPTHIRHQSGSYG 377
Query: 121 EALAVHKKKFQNDKDNTERLQQWKEALSQAANLSGQHYK-HG 161
E L H+++FQN++ N ERL+QWK AL QAANLSG HY HG
Sbjct: 378 EHLTKHEERFQNNEKNMERLRQWKIALIQAANLSGYHYSPHG 503
Score = 42.0 bits (97), Expect = 6e-04
Identities = 20/43 (46%), Positives = 28/43 (64%)
Frame = +2
Query: 504 GTRKIKMINMKFPSMESDIDWNGNAFEKMTNLKTFITENGHHS 546
GT I+MI ++F S + +W+G A +KMTNLKT I E + S
Sbjct: 503 GTGNIEMIYLEFDSTARETEWDGMACKKMTNLKTLIIEYANFS 631
>BF648630 weakly similar to GP|18033111|g functional candidate resistance
protein KR1 {Glycine max}, partial (13%)
Length = 654
Score = 228 bits (582), Expect = 4e-60
Identities = 122/208 (58%), Positives = 148/208 (70%), Gaps = 6/208 (2%)
Frame = +1
Query: 302 GPGSRVIITTRNKHLLKIHGIESTHAVEGLNATEALELLRWMAFKENV-PSSHEDILNRA 360
G GSRVIITTR+KHLL HGI + GLN +ALELLR AFK SS++ ILNRA
Sbjct: 4 GRGSRVIITTRDKHLLSSHGITKIYEAYGLNKEQALELLRTKAFKSKKNDSSYDYILNRA 183
Query: 361 LTYASGLPLAIVIIGSNLVGRSVQDSMSTLDGYEEIPNKEIQRILKVSYDSLEKEEQSVF 420
+ YASGLPLA+ ++GSNL G S + STLD YE IP ++IQ+ILKVS+D+L++E+QSVF
Sbjct: 184 IKYASGLPLALEVVGSNLFGMSTTECESTLDKYERIPPEDIQKILKVSFDALDEEQQSVF 363
Query: 421 LDIACCFKGCKWPEVKEILHAHYGHCIVHHVAVLAEKSLMD-----HLKYDSYVTLHDLI 475
LDIAC F C+ V+EIL HYGHCI H+ L +KSL+ H VTLHDL+
Sbjct: 364 LDIACFFNWCESAYVEEILEYHYGHCIKSHLRALVDKSLIKTSIQRHGMKFELVTLHDLL 543
Query: 476 EDMGKEVVRQESPDEPGERSRLWFERDI 503
EDMGKE+VR ES EPGERSRLW+ DI
Sbjct: 544 EDMGKEIVRHESIKEPGERSRLWYHDDI 627
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.318 0.136 0.400
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 17,904,034
Number of Sequences: 36976
Number of extensions: 246659
Number of successful extensions: 1770
Number of sequences better than 10.0: 217
Number of HSP's better than 10.0 without gapping: 1588
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1609
length of query: 576
length of database: 9,014,727
effective HSP length: 101
effective length of query: 475
effective length of database: 5,280,151
effective search space: 2508071725
effective search space used: 2508071725
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 61 (28.1 bits)
Medicago: description of AC138133.14


