

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC138133.11 - phase: 0
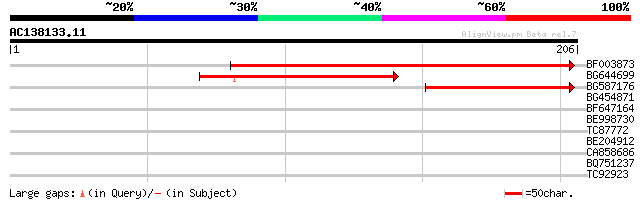
(206 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BF003873 similar to GP|14715222|em putative polyprotein {Cicer a... 224 2e-59
BG644699 similar to PIR|T07863|T078 probable polyprotein - pinea... 64 3e-11
BG587176 weakly similar to PIR|G84493|G84 probable retroelement ... 57 5e-09
BG454871 weakly similar to GP|10140673|g putative gag-pol polypr... 33 0.012
BF647164 weakly similar to GP|6691191|gb F7F22.15 {Arabidopsis t... 30 0.65
BE998730 29 1.4
TC87772 weakly similar to GP|8574455|gb|AAF77578.1| pepper ester... 29 1.4
BE204912 similar to PIR|T02686|T026 probable Ser/Thr protein kin... 27 4.2
CA858686 homologue to GP|8489871|gb| DEAD box protein P68 {Pisum... 27 4.2
BQ751237 weakly similar to GP|16197477|dbj ORF1 {TT virus}, part... 27 7.2
TC92923 similar to GP|6466937|gb|AAF13073.1| putative retroeleme... 27 7.2
>BF003873 similar to GP|14715222|em putative polyprotein {Cicer arietinum},
partial (82%)
Length = 559
Score = 224 bits (570), Expect = 2e-59
Identities = 110/125 (88%), Positives = 115/125 (92%)
Frame = +2
Query: 81 GVGRALKSRKLTPKFIGPYQISERIGTVAYRVGLPPHLSNLHDVFHVSQLRKYVADPSHV 140
GVGRALKS+KLT +FIGPYQISER+GTVAYRVGLPPHL NLHDVFHVSQLRKYV DPSHV
Sbjct: 2 GVGRALKSKKLTVRFIGPYQISERVGTVAYRVGLPPHLLNLHDVFHVSQLRKYVPDPSHV 181
Query: 141 IPRDDVQVRDNLTVETLPLRIDDRKVKALRGKEIPLVRVVWGGATGESLTWELESKMRDS 200
I DDVQVRDNLTVETLP+RIDDRKVK LRGKEIPLVRVVW A GESLTWELESKM +S
Sbjct: 182 IQSDDVQVRDNLTVETLPVRIDDRKVKTLRGKEIPLVRVVWDRANGESLTWELESKMVES 361
Query: 201 YPELF 205
YPELF
Sbjct: 362 YPELF 376
>BG644699 similar to PIR|T07863|T078 probable polyprotein - pineapple
retrotransposon dea1 (fragment), partial (5%)
Length = 231
Score = 64.3 bits (155), Expect = 3e-11
Identities = 32/73 (43%), Positives = 48/73 (64%), Gaps = 1/73 (1%)
Frame = +2
Query: 70 DHVFLRVTPMT-GVGRALKSRKLTPKFIGPYQISERIGTVAYRVGLPPHLSNLHDVFHVS 128
+ V L+V P G R K KL+ ++IGP+++ +RIG VAY + LPP LS +H VFHVS
Sbjct: 2 EQVLLKVLPTERGDCRFGKRGKLSLRYIGPFEVIKRIGEVAYELALPPGLSGVHPVFHVS 181
Query: 129 QLRKYVADPSHVI 141
++Y D +++I
Sbjct: 182 MFKRYHGDGNYII 220
>BG587176 weakly similar to PIR|G84493|G84 probable retroelement pol
polyprotein [imported] - Arabidopsis thaliana, partial
(1%)
Length = 729
Score = 57.0 bits (136), Expect = 5e-09
Identities = 26/54 (48%), Positives = 37/54 (68%)
Frame = -1
Query: 152 LTVETLPLRIDDRKVKALRGKEIPLVRVVWGGATGESLTWELESKMRDSYPELF 205
L +ET P+RI DR KA+R K I +V++VW + E +TWE E++M+ YPE F
Sbjct: 717 LDLETRPVRILDRMEKAMRKKPIQMVKIVWDCSGREEITWETEARMKADYPEWF 556
>BG454871 weakly similar to GP|10140673|g putative gag-pol polyprotein {Oryza
sativa (japonica cultivar-group)}, partial (7%)
Length = 674
Score = 32.7 bits (73), Expect(2) = 0.012
Identities = 21/73 (28%), Positives = 34/73 (45%), Gaps = 2/73 (2%)
Frame = +1
Query: 49 SQSRQKSYHDKRRKALEFQEGDHVFLRVTPMTGVGRALKSRKL--TPKFIGPYQISERIG 106
+Q K DK+R+ EFQ G+HV +++ P AL+ + +P F +
Sbjct: 388 AQQTMKHQADKKRRHFEFQLGEHVLVKLQPYQQSSVALRKYQKFGSPNFGSLLTVCSL*V 567
Query: 107 TVAYRVGLPPHLS 119
A+ PP+LS
Sbjct: 568 ESAFHCKSPPYLS 606
Score = 21.9 bits (45), Expect(2) = 0.012
Identities = 8/10 (80%), Positives = 9/10 (90%)
Frame = +2
Query: 1 MAPFEALYGR 10
M PF+ALYGR
Sbjct: 245 MTPFKALYGR 274
>BF647164 weakly similar to GP|6691191|gb F7F22.15 {Arabidopsis thaliana},
partial (6%)
Length = 469
Score = 30.0 bits (66), Expect = 0.65
Identities = 21/69 (30%), Positives = 33/69 (47%)
Frame = +1
Query: 44 EKMKASQSRQKSYHDKRRKALEFQEGDHVFLRVTPMTGVGRALKSRKLTPKFIGPYQISE 103
E K + R K +HD+R EF+EG+ V L + + L KL + GP+Q+
Sbjct: 142 ENAKIYKERTKKWHDRRIIRREFREGELVLLFNSRL-----KLFPGKLRSHWSGPFQVKN 306
Query: 104 RIGTVAYRV 112
+ + A V
Sbjct: 307 VMPSGAVEV 333
>BE998730
Length = 820
Score = 28.9 bits (63), Expect = 1.4
Identities = 16/46 (34%), Positives = 21/46 (44%), Gaps = 6/46 (13%)
Frame = +1
Query: 167 KALRGKEIPLVRVV------WGGATGESLTWELESKMRDSYPELFV 206
K RGK+ P+ + WG SL W + K+ SYP L V
Sbjct: 592 KQTRGKQFPVRHTLFNMFKPWGVFVQNSLHWVMAKKLDGSYPWLIV 729
>TC87772 weakly similar to GP|8574455|gb|AAF77578.1| pepper esterase
{Capsicum annuum}, partial (34%)
Length = 1313
Score = 28.9 bits (63), Expect = 1.4
Identities = 16/43 (37%), Positives = 25/43 (57%), Gaps = 4/43 (9%)
Frame = -2
Query: 117 HLSNLHDVFHVSQLRKYVADPS----HVIPRDDVQVRDNLTVE 155
H+SN HDV + LR ++ S H +P D V V+D++ V+
Sbjct: 628 HVSNQHDVLAMDLLRF*SSEESPRSLHRLPEDGVPVQDDIRVK 500
>BE204912 similar to PIR|T02686|T026 probable Ser/Thr protein kinase
[imported] - Arabidopsis thaliana, partial (2%)
Length = 603
Score = 27.3 bits (59), Expect = 4.2
Identities = 18/47 (38%), Positives = 26/47 (55%), Gaps = 2/47 (4%)
Frame = +3
Query: 108 VAYRVGLPPHLS--NLHDVFHVSQLRKYVADPSHVIPRDDVQVRDNL 152
VAYR P LS N+ D+F++S LR +A S++ D + D L
Sbjct: 186 VAYRAQSPNFLSLSNISDIFNLSPLR--IAKASNIEAEDKKLIPDQL 320
>CA858686 homologue to GP|8489871|gb| DEAD box protein P68 {Pisum sativum},
partial (31%)
Length = 598
Score = 27.3 bits (59), Expect = 4.2
Identities = 24/92 (26%), Positives = 37/92 (40%), Gaps = 2/92 (2%)
Frame = +2
Query: 47 KASQSRQKSYHDKRRKALEFQEGDHVFLRVTPMTGVGRA--LKSRKLTPKFIGPYQISER 104
++ R+ + HD R + V R +TGV L K T +I + R
Sbjct: 308 RSQSEREAALHDFRSSSTSILVATDVASRGLDVTGVSHVINLDLPKTTEDYIHRIGRTGR 487
Query: 105 IGTVAYRVGLPPHLSNLHDVFHVSQLRKYVAD 136
G+ G+ D+F VS +RK +AD
Sbjct: 488 AGST----GIATSFYTDRDMFLVSNIRKAIAD 571
>BQ751237 weakly similar to GP|16197477|dbj ORF1 {TT virus}, partial (3%)
Length = 681
Score = 26.6 bits (57), Expect = 7.2
Identities = 14/29 (48%), Positives = 19/29 (65%)
Frame = +1
Query: 159 LRIDDRKVKALRGKEIPLVRVVWGGATGE 187
LR+DD V A+ G+E+ VRV GG G+
Sbjct: 577 LRVDDVDVGAVDGRELVAVRV--GGGAGD 657
>TC92923 similar to GP|6466937|gb|AAF13073.1| putative retroelement pol
polyprotein {Arabidopsis thaliana}, partial (1%)
Length = 625
Score = 26.6 bits (57), Expect = 7.2
Identities = 10/19 (52%), Positives = 14/19 (73%)
Frame = +3
Query: 119 SNLHDVFHVSQLRKYVADP 137
S +H +FHVSQL++ V P
Sbjct: 228 STIHSIFHVSQLKRAVWGP 284
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.320 0.138 0.410
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,753,001
Number of Sequences: 36976
Number of extensions: 64721
Number of successful extensions: 376
Number of sequences better than 10.0: 22
Number of HSP's better than 10.0 without gapping: 374
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 376
length of query: 206
length of database: 9,014,727
effective HSP length: 92
effective length of query: 114
effective length of database: 5,612,935
effective search space: 639874590
effective search space used: 639874590
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 56 (26.2 bits)
Medicago: description of AC138133.11


