

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
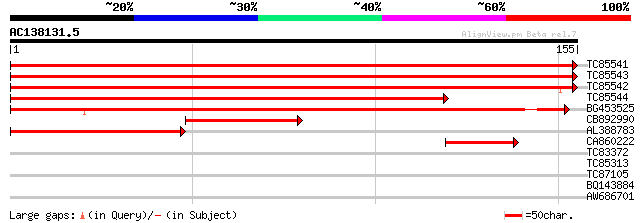
Query= AC138131.5 + phase: 0 /partial
(155 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC85541 homologue to GP|166867|gb|AAA32866.1|| ribosomal protein... 321 6e-89
TC85543 homologue to GP|166867|gb|AAA32866.1|| ribosomal protein... 315 4e-87
TC85542 homologue to SP|P17093|RS11_SOYBN 40S ribosomal protein ... 301 5e-83
TC85544 homologue to GP|6984142|gb|AAF34771.1| 40S ribosomal pro... 241 7e-65
BG453525 similar to SP|P79013|RS11 40S ribosomal protein S11. [F... 208 6e-55
CB892990 similar to PIR|D35542|D355 ribosomal protein S11 - soyb... 55 9e-09
AL388783 similar to SP|P79013|RS11_ 40S ribosomal protein S11. [... 46 5e-06
CA860222 similar to PIR|S41784|S417 ribosomal protein S11.e cyt... 41 2e-04
TC83372 weakly similar to GP|20339360|gb|AAM19353.1 TIR-similar-... 30 0.51
TC85313 PIR|T09338|T09338 DnaJ-like protein MsJ1 - alfalfa, comp... 28 1.1
TC87105 similar to PIR|T14329|T14329 dermal glycoprotein precurs... 28 1.9
BQ143884 similar to GP|163002|gb|AA elastin {Bos taurus}, partia... 27 4.3
AW686701 similar to GP|21553679|gb| putative myo-inositol monoph... 25 9.6
>TC85541 homologue to GP|166867|gb|AAA32866.1|| ribosomal protein S11
(probable start codon at bp 67) {Arabidopsis thaliana},
partial (87%)
Length = 883
Score = 321 bits (823), Expect = 6e-89
Identities = 155/155 (100%), Positives = 155/155 (100%)
Frame = +3
Query: 1 TEKAFLKQPKVFLCSKKSGKGKRPGKGGNRFFKSVGLGFKTPREAIDGTYIDKKCPFTGN 60
TEKAFLKQPKVFLCSKKSGKGKRPGKGGNRFFKSVGLGFKTPREAIDGTYIDKKCPFTGN
Sbjct: 123 TEKAFLKQPKVFLCSKKSGKGKRPGKGGNRFFKSVGLGFKTPREAIDGTYIDKKCPFTGN 302
Query: 61 VSIRGRILAGTCHSAKMNRTIVVRRNYLHFIKKYQRYEKRHSNIPAHVSPCFRVKEGDHV 120
VSIRGRILAGTCHSAKMNRTIVVRRNYLHFIKKYQRYEKRHSNIPAHVSPCFRVKEGDHV
Sbjct: 303 VSIRGRILAGTCHSAKMNRTIVVRRNYLHFIKKYQRYEKRHSNIPAHVSPCFRVKEGDHV 482
Query: 121 IIGQCRPLSKTVRFNVLKVIPAGSSSGAGKKAFTG 155
IIGQCRPLSKTVRFNVLKVIPAGSSSGAGKKAFTG
Sbjct: 483 IIGQCRPLSKTVRFNVLKVIPAGSSSGAGKKAFTG 587
>TC85543 homologue to GP|166867|gb|AAA32866.1|| ribosomal protein S11
(probable start codon at bp 67) {Arabidopsis thaliana},
partial (87%)
Length = 802
Score = 315 bits (808), Expect = 4e-87
Identities = 151/155 (97%), Positives = 154/155 (98%)
Frame = +2
Query: 1 TEKAFLKQPKVFLCSKKSGKGKRPGKGGNRFFKSVGLGFKTPREAIDGTYIDKKCPFTGN 60
TEKAFLKQPKVFLCSKKSGKGKRPGKGGNRF+KSVGLGFKTPREA +GTYIDKKCPFTGN
Sbjct: 92 TEKAFLKQPKVFLCSKKSGKGKRPGKGGNRFWKSVGLGFKTPREATEGTYIDKKCPFTGN 271
Query: 61 VSIRGRILAGTCHSAKMNRTIVVRRNYLHFIKKYQRYEKRHSNIPAHVSPCFRVKEGDHV 120
VSIRGRILAGTCHSAKMNRTI+VRRNYLHFIKKYQRYEKRHSNIPAHVSPCFRVKEGDHV
Sbjct: 272 VSIRGRILAGTCHSAKMNRTIIVRRNYLHFIKKYQRYEKRHSNIPAHVSPCFRVKEGDHV 451
Query: 121 IIGQCRPLSKTVRFNVLKVIPAGSSSGAGKKAFTG 155
IIGQCRPLSKTVRFNVLKVIPAGSSSGAGKKAFTG
Sbjct: 452 IIGQCRPLSKTVRFNVLKVIPAGSSSGAGKKAFTG 556
>TC85542 homologue to SP|P17093|RS11_SOYBN 40S ribosomal protein S11.
[Soybean] {Glycine max}, complete
Length = 781
Score = 301 bits (772), Expect = 5e-83
Identities = 147/156 (94%), Positives = 152/156 (97%), Gaps = 1/156 (0%)
Frame = +2
Query: 1 TEKAFLKQPKVFLCSKKSGKGKRPGKGGNRFFKSVGLGFKTPREAIDGTYIDKKCPFTGN 60
TEKAFLKQ KVFLCSK SGKGKRPGKGGNRF K+VGLGFKTPREAI+GTYIDKKCPFTGN
Sbjct: 95 TEKAFLKQNKVFLCSKISGKGKRPGKGGNRFHKTVGLGFKTPREAIEGTYIDKKCPFTGN 274
Query: 61 VSIRGRILAGTCHSAKMNRTIVVRRNYLHFIKKYQRYEKRHSNIPAHVSPCFRVKEGDHV 120
VSIRGRI++GTCHSAKMN+TIVVRRNYLHFIKKYQRYEKRHSNIPAHVSPCFRVKEGDHV
Sbjct: 275 VSIRGRIISGTCHSAKMNKTIVVRRNYLHFIKKYQRYEKRHSNIPAHVSPCFRVKEGDHV 454
Query: 121 IIGQCRPLSKTVRFNVLKVIPAGSSSGAG-KKAFTG 155
IIGQCRPLSKTVRFNVLKVIPAGSSSGAG KKAFTG
Sbjct: 455 IIGQCRPLSKTVRFNVLKVIPAGSSSGAGAKKAFTG 562
>TC85544 homologue to GP|6984142|gb|AAF34771.1| 40S ribosomal protein S11
{Euphorbia esula}, partial (76%)
Length = 1335
Score = 241 bits (616), Expect = 7e-65
Identities = 114/120 (95%), Positives = 117/120 (97%)
Frame = +1
Query: 1 TEKAFLKQPKVFLCSKKSGKGKRPGKGGNRFFKSVGLGFKTPREAIDGTYIDKKCPFTGN 60
TEKAFLKQPKVFLCSKKSGKGKRPGKGGNRFFKSVGLGF TPREAIDGTYIDKKCPFTGN
Sbjct: 28 TEKAFLKQPKVFLCSKKSGKGKRPGKGGNRFFKSVGLGFNTPREAIDGTYIDKKCPFTGN 207
Query: 61 VSIRGRILAGTCHSAKMNRTIVVRRNYLHFIKKYQRYEKRHSNIPAHVSPCFRVKEGDHV 120
VSI+GRILAGTCHSAKMN TIVVRRNYLHFIKKYQRYEKRHSNIPAHVSPCFRVKEG ++
Sbjct: 208 VSIQGRILAGTCHSAKMNTTIVVRRNYLHFIKKYQRYEKRHSNIPAHVSPCFRVKEGSNM 387
>BG453525 similar to SP|P79013|RS11 40S ribosomal protein S11. [Fission
yeast] {Schizosaccharomyces pombe}, partial (90%)
Length = 638
Score = 208 bits (530), Expect = 6e-55
Identities = 103/154 (66%), Positives = 121/154 (77%), Gaps = 1/154 (0%)
Frame = +3
Query: 1 TEKAFLKQPKVFLCSKKSG-KGKRPGKGGNRFFKSVGLGFKTPREAIDGTYIDKKCPFTG 59
+E+AF KQP VF +KK G K KR GKGG R+ K VGLGF+TP+ AI+G YIDKKCPFTG
Sbjct: 108 SERAFQKQPHVFQNTKKQGGKTKRAGKGGRRWVKDVGLGFRTPKTAIEGNYIDKKCPFTG 287
Query: 60 NVSIRGRILAGTCHSAKMNRTIVVRRNYLHFIKKYQRYEKRHSNIPAHVSPCFRVKEGDH 119
VSIRGRIL GT S KM+RT+V+RR YLHF+ KY RYE+RH N+ AHVSP FRV+EGD
Sbjct: 288 LVSIRGRILTGTVVSTKMHRTLVIRREYLHFVPKYSRYERRHKNLSAHVSPAFRVEEGDQ 467
Query: 120 VIIGQCRPLSKTVRFNVLKVIPAGSSSGAGKKAF 153
V +GQCRPLSKTVRFNVL+V+P +G KAF
Sbjct: 468 VTVGQCRPLSKTVRFNVLRVLP---RTGKAVKAF 560
>CB892990 similar to PIR|D35542|D355 ribosomal protein S11 - soybean
(fragment), partial (22%)
Length = 127
Score = 55.5 bits (132), Expect = 9e-09
Identities = 26/32 (81%), Positives = 27/32 (84%)
Frame = +1
Query: 49 TYIDKKCPFTGNVSIRGRILAGTCHSAKMNRT 80
TYIDKK P TG VSIRGR+ AGTCHSAK NRT
Sbjct: 31 TYIDKKGPCTGKVSIRGRMSAGTCHSAKKNRT 126
>AL388783 similar to SP|P79013|RS11_ 40S ribosomal protein S11. [Fission
yeast] {Schizosaccharomyces pombe}, partial (20%)
Length = 542
Score = 46.2 bits (108), Expect = 5e-06
Identities = 22/48 (45%), Positives = 31/48 (63%)
Frame = +3
Query: 1 TEKAFLKQPKVFLCSKKSGKGKRPGKGGNRFFKSVGLGFKTPREAIDG 48
+E+AF KQP +F +K K + K R++K VGLGFKTP+E + G
Sbjct: 147 SERAFQKQPGIFTSAKVITKKGKSKKKNTRWYKDVGLGFKTPKEVM*G 290
>CA860222 similar to PIR|S41784|S417 ribosomal protein S11.e cytosolic -
yeast (Saccharomyces cerevisiae), partial (21%)
Length = 298
Score = 40.8 bits (94), Expect = 2e-04
Identities = 18/20 (90%), Positives = 19/20 (95%)
Frame = +2
Query: 120 VIIGQCRPLSKTVRFNVLKV 139
V +GQCRPLSKTVRFNVLKV
Sbjct: 2 VTVGQCRPLSKTVRFNVLKV 61
>TC83372 weakly similar to GP|20339360|gb|AAM19353.1
TIR-similar-domain-containing protein TSDC {Pisum
sativum}, partial (21%)
Length = 1151
Score = 29.6 bits (65), Expect = 0.51
Identities = 13/45 (28%), Positives = 21/45 (45%)
Frame = -3
Query: 67 ILAGTCHSAKMNRTIVVRRNYLHFIKKYQRYEKRHSNIPAHVSPC 111
IL TC + + R YL +IK ++ + H N+ H+ C
Sbjct: 222 ILLATCGQKDLASITITRNAYLFWIKIFRNIIQDHKNLAIHIC*C 88
>TC85313 PIR|T09338|T09338 DnaJ-like protein MsJ1 - alfalfa, complete
Length = 1927
Score = 28.5 bits (62), Expect = 1.1
Identities = 16/34 (47%), Positives = 17/34 (49%)
Frame = +1
Query: 13 LCSKKSGKGKRPGKGGNRFFKSVGLGFKTPREAI 46
LCSK GKG + G G R F G G K R I
Sbjct: 724 LCSKCKGKGSKSGTAG-RCFGCQGTGMKITRRQI 822
>TC87105 similar to PIR|T14329|T14329 dermal glycoprotein precursor
extracellular - carrot (fragment), partial (39%)
Length = 1540
Score = 27.7 bits (60), Expect = 1.9
Identities = 9/32 (28%), Positives = 18/32 (56%)
Frame = -1
Query: 72 CHSAKMNRTIVVRRNYLHFIKKYQRYEKRHSN 103
CH +NR + + +Y+ + K+ R+ HS+
Sbjct: 313 CHCNHVNRRVSIEMSYIGLVLKWMRHSSCHSS 218
>BQ143884 similar to GP|163002|gb|AA elastin {Bos taurus}, partial (2%)
Length = 1024
Score = 26.6 bits (57), Expect = 4.3
Identities = 15/36 (41%), Positives = 21/36 (57%)
Frame = -3
Query: 40 KTPREAIDGTYIDKKCPFTGNVSIRGRILAGTCHSA 75
KT REA+ + TGN++IR + +GTCH A
Sbjct: 527 KTSREALRAAN-EYHENHTGNMNIRKTVDSGTCHRA 423
>AW686701 similar to GP|21553679|gb| putative myo-inositol monophosphatase
{Arabidopsis thaliana}, partial (35%)
Length = 408
Score = 25.4 bits (54), Expect = 9.6
Identities = 12/38 (31%), Positives = 18/38 (46%)
Frame = -2
Query: 56 PFTGNVSIRGRILAGTCHSAKMNRTIVVRRNYLHFIKK 93
P+T S I + TC+S K+ T N H +K+
Sbjct: 269 PWTTKSSFIEHIQSSTCNSTKIQSTGATHSNRSHLVKQ 156
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.322 0.139 0.424
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,974,117
Number of Sequences: 36976
Number of extensions: 69079
Number of successful extensions: 371
Number of sequences better than 10.0: 27
Number of HSP's better than 10.0 without gapping: 371
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 371
length of query: 155
length of database: 9,014,727
effective HSP length: 88
effective length of query: 67
effective length of database: 5,760,839
effective search space: 385976213
effective search space used: 385976213
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 54 (25.4 bits)
Medicago: description of AC138131.5


