

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC138131.4 + phase: 0 /pseudo
(1189 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
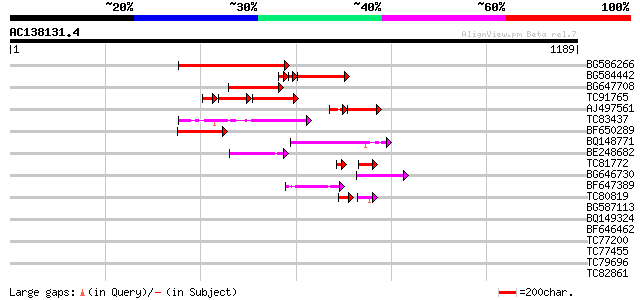
Score E
Sequences producing significant alignments: (bits) Value
BG586266 similar to GP|7267666|em RNA-directed DNA polymerase-li... 208 1e-53
BG584442 114 1e-27
BG647708 weakly similar to GP|13786450|gb| putative reverse tran... 112 1e-24
TC91765 weakly similar to GP|19881779|gb|AAM01180.1 Putative ret... 90 2e-23
AJ497561 weakly similar to GP|10140689|gb putative non-LTR retro... 76 6e-19
TC83437 weakly similar to PIR|D86384|D86384 unknown protein [imp... 87 3e-17
BF650289 weakly similar to GP|9049283|dbj| orf129a {Beta vulgari... 80 6e-15
BQ148771 71 3e-12
BE248682 similar to GP|18568269|gb putative gag-pol polyprotein ... 70 6e-12
TC81772 58 4e-10
BG646730 60 5e-09
BF647389 44 4e-04
TC80819 weakly similar to GP|10140689|gb|AAG13524.1 putative non... 37 7e-04
BG587113 weakly similar to PIR|A84888|A8 hypothetical protein At... 40 0.005
BQ149324 37 0.045
BF646462 34 0.38
TC77200 similar to GP|22597162|gb|AAN03468.1 bZIP transcription ... 30 5.5
TC77455 similar to GP|22335695|dbj|BAC10549. nine-cis-epoxycarot... 30 7.2
TC79696 similar to GP|15216028|emb|CAC51424. amino acid permease... 29 9.5
TC82861 similar to GP|6630450|gb|AAF19538.1| F23N19.10 {Arabidop... 29 9.5
>BG586266 similar to GP|7267666|em RNA-directed DNA polymerase-like protein
{Arabidopsis thaliana}, partial (18%)
Length = 789
Score = 208 bits (529), Expect = 1e-53
Identities = 106/234 (45%), Positives = 147/234 (62%)
Frame = -3
Query: 354 RLKVVLDKCISEFQSAFVPGRSIFDNVMAAIEIIHHMKIKVKGNVGEVALKLDISKAYDR 413
R++ +L IS QSAFVPGR+I DNV+ +I+H+++ +A+K D++KAYDR
Sbjct: 709 RMQPLLRSIISPSQSAFVPGRAISDNVLITHKILHYLRQSGAKKHVSMAVKTDMTKAYDR 530
Query: 414 IEWNYLRQVMDKMGFSEQWIKWIMSCVETVDYFVLVNGNVTGPIIPGRGLRQGDPLSPYL 473
I WN+LR+V+ ++GF WI WIM CV TV Y L+NG G ++P RGLRQGDPLSPYL
Sbjct: 529 IAWNFLREVLTRLGFHGIWISWIMECVSTVSYSFLINGGPQGRVLPSRGLRQGDPLSPYL 350
Query: 474 FIICAEGLSALIRKAEARGDINGVKVCNNAPIISHLLFADDCFLFFRANLNKAQKMRNIL 533
FI+C E LS L ++A +G + GVKV N P I+HLLFADD F ++N + + +I+
Sbjct: 349 FILCTEVLSGLCQQALRKGTLPGVKVARNCPPINHLLFADDTMFFGKSNASSCAILLSIM 170
Query: 534 SVYEKASGQAVNLQKSEFFCSRNVSPADHKNIANILGVKTVLGTGKYLGLPSMI 587
Y ASG+ +N KS S S A + L + GTGKYLG +++
Sbjct: 169 DKYRAASGRCIN*TKSAITFSSKTSQAIIDRVKGELKIAKEGGTGKYLGYRNIL 8
>BG584442
Length = 775
Score = 114 bits (286), Expect(3) = 1e-27
Identities = 60/108 (55%), Positives = 73/108 (67%)
Frame = +1
Query: 604 KKINSWSSKCLSKAGREVLIKFVLQSIPTYFMSLFTLPLSLCDEIEKMMNSFWWGHSGSN 663
KKIN +KCLSK EV+IK+ LQSI +Y MS+F L S DEIEK+MN+F W H G N
Sbjct: 379 KKINF*RNKCLSKVM*EVMIKYALQSISSYVMSIFLLLNSQVDEIEKIMNTFSWVHVGEN 558
Query: 664 KKGIHWMSWDKLSMHKKDGGMSFKNLGTFNLAMLGKQGWRILTQSDTL 711
+KG+HWMS +KL +HK GGM F + TFN+ MLGKQ L TL
Sbjct: 559 RKGMHWMS*EKLFVHKNYGGMGFTDFTTFNIPMLGKQV*SFLLNRTTL 702
Score = 24.6 bits (52), Expect(3) = 1e-27
Identities = 9/17 (52%), Positives = 13/17 (75%)
Frame = +3
Query: 586 MIGRSKKATFNFIKDRV 602
M+ S KATF++IKD +
Sbjct: 324 MLSTSNKATFSYIKDEI 374
Score = 23.5 bits (49), Expect(3) = 1e-27
Identities = 11/19 (57%), Positives = 12/19 (62%)
Frame = +2
Query: 565 IANILGVKTVLGTGKYLGL 583
I IL V+ VLG KY GL
Sbjct: 239 ITYILDVRAVLGRDKYFGL 295
>BG647708 weakly similar to GP|13786450|gb| putative reverse transcriptase
{Oryza sativa}, partial (9%)
Length = 708
Score = 112 bits (279), Expect = 1e-24
Identities = 55/115 (47%), Positives = 78/115 (67%)
Frame = +1
Query: 459 PGRGLRQGDPLSPYLFIICAEGLSALIRKAEARGDINGVKVCNNAPIISHLLFADDCFLF 518
P +GLRQGDPLSPYLFI+CA LS L+++ + +++G++V + P I+HLLFADD LF
Sbjct: 4 PEKGLRQGDPLSPYLFILCANVLSGLLKREGNKQNLHGIQVARSDPKITHLLFADDSLLF 183
Query: 519 FRANLNKAQKMRNILSVYEKASGQAVNLQKSEFFCSRNVSPADHKNIANILGVKT 573
RANL +A + +L Y+ ASGQ VN +KSE S+NV + + I + +KT
Sbjct: 184 ARANLTEAATIMQVLHSYQSASGQLVNFEKSEVSYSQNVPNQEKEMICQQIAIKT 348
>TC91765 weakly similar to GP|19881779|gb|AAM01180.1 Putative retroelement
{Oryza sativa (japonica cultivar-group)}, partial (1%)
Length = 625
Score = 89.7 bits (221), Expect = 6e-18
Identities = 49/97 (50%), Positives = 64/97 (65%)
Frame = +3
Query: 509 LLFADDCFLFFRANLNKAQKMRNILSVYEKASGQAVNLQKSEFFCSRNVSPADHKNIANI 568
LLF R + AQ M+NIL +YE+ SG+A++L+KS +CSRNV +I I
Sbjct: 279 LLFEGAPLRSLRVEEHHAQIMKNILILYEEDSGKAISLRKS*IYCSRNVPDILKTSITYI 458
Query: 569 LGVKTVLGTGKYLGLPSMIGRSKKATFNFIKDRVWKK 605
LGV+ +LGT KYLGLPSMIGR + TF+ IK VW+K
Sbjct: 459 LGVQFMLGTCKYLGLPSMIGRDRTTTFSSIKGGVWQK 569
Score = 86.7 bits (213), Expect(2) = 2e-23
Identities = 41/69 (59%), Positives = 50/69 (72%)
Frame = +2
Query: 439 CVETVDYFVLVNGNVTGPIIPGRGLRQGDPLSPYLFIICAEGLSALIRKAEARGDINGVK 498
CVE+ DY+VLVN + PIIP RGL+QGD LSPY+FIIC EGLS LI A+ RGD +G
Sbjct: 104 CVESNDYYVLVNNDAVDPIIPSRGLQQGDHLSPYIFIICVEGLSFLIPHAKERGDTHGTS 283
Query: 499 VCNNAPIIS 507
+ AP +S
Sbjct: 284 I*RGAPPVS 310
Score = 42.4 bits (98), Expect(2) = 2e-23
Identities = 16/32 (50%), Positives = 26/32 (81%)
Frame = +1
Query: 405 LDISKAYDRIEWNYLRQVMDKMGFSEQWIKWI 436
L ISK Y+R++ +YL+++M KMGF+ +WI W+
Sbjct: 1 LYISKVYNRVD*DYLKEIMIKMGFNNRWIYWM 96
Score = 35.4 bits (80), Expect = 0.13
Identities = 17/29 (58%), Positives = 20/29 (68%)
Frame = +1
Query: 595 FNFIKDRVWKKINSWSSKCLSKAGREVLI 623
F ++ KKINSWS CLSKA REV+I
Sbjct: 538 FLLLRVECGKKINSWSDICLSKACREVMI 624
>AJ497561 weakly similar to GP|10140689|gb putative non-LTR retroelement
reverse transcriptase {Oryza sativa (japonica
cultivar-group)}, partial (2%)
Length = 621
Score = 76.3 bits (186), Expect(2) = 6e-19
Identities = 33/72 (45%), Positives = 50/72 (68%)
Frame = +2
Query: 708 SDTLIARIYKARYFPRCNFFESELGHNPSFVWRSICKSKFILKAGSRWKIGDGTSISVWN 767
S +L+++I K +YFP+ +F + LGHNPSF WRS+ ++ +L G RW IGDG+ I+V +
Sbjct: 404 STSLLSKILKFKYFPQWDFSYANLGHNPSFTWRSLLSTQSLLTLGHRWMIGDGSQINVSS 583
Query: 768 NYWMRNDVTIFP 779
W+RN T+ P
Sbjct: 584 MSWIRNRPTLRP 619
Score = 37.4 bits (85), Expect(2) = 6e-19
Identities = 16/36 (44%), Positives = 26/36 (71%)
Frame = +3
Query: 672 WDKLSMHKKDGGMSFKNLGTFNLAMLGKQGWRILTQ 707
++ L+ HK GG+ +N FN++MLGKQ W++L+Q
Sbjct: 303 FNNLARHKSVGGLGLQNQ-EFNISMLGKQRWKLLSQ 407
>TC83437 weakly similar to PIR|D86384|D86384 unknown protein [imported] -
Arabidopsis thaliana, partial (6%)
Length = 951
Score = 87.4 bits (215), Expect = 3e-17
Identities = 84/292 (28%), Positives = 129/292 (43%), Gaps = 12/292 (4%)
Frame = +2
Query: 354 RLKVVLDKCISEFQSAFVPGRSIFDNVMAAIEIIHHMKIKVKGNVGEVALKLDISKAYDR 413
RL+VV+ IS+ QSAFV R I + V + M++ ++ + K +
Sbjct: 170 RLRVVIGSVISDAQSAFVKNRQILEMVF-----L*QMRLWMRLR--------N*RKIFCC 310
Query: 414 IEWNYLRQVMDKMG-----------FSEQWIKWIMSCVETVDYFVLVNGNVTGPIIPGRG 462
+ W R + +G F W KWI CV T VLVNG+ T ++ +
Sbjct: 311 LRWILKRLITLSIGLIWILF*VGMSFLVLWRKWIKECVSTATTSVLVNGSPTNVLM--KS 484
Query: 463 LRQGDPLSPYLFIICAEGLSALIRKAEARGDINGVKVCNNAPIISHLLFADDCFLFFRAN 522
L Q + Y F G +N V ++SHL FA+D L N
Sbjct: 485 LVQTQLFTRYSF-----------------GVVNPV-------VVSHLQFANDTLLLETKN 592
Query: 523 LNKAQKMRNILSVYEKASGQAVNLQKSEFFCSRNVSPADHKNIANILGVKTVLGTGKYLG 582
+ +R L ++ SG VN KS C N++P+ A++L K YLG
Sbjct: 593 WANIRALRAALVIF*AMSGLKVNFHKSGLVCV-NIAPSWLSEAASVLSWKVGKVPFLYLG 769
Query: 583 LPSMIGRSKKATF-NFIKDRVWKKINSWSSKCLSKAGREVLIKFVLQSIPTY 633
+P + G S++ +F I +R+ ++ W+S+ LS GR VL+K VL S+ Y
Sbjct: 770 MP-IEGNSRRLSFWEPIVNRIKARLTGWNSRFLSFGGRLVLLKSVLTSLSVY 922
>BF650289 weakly similar to GP|9049283|dbj| orf129a {Beta vulgaris}, partial
(69%)
Length = 616
Score = 79.7 bits (195), Expect = 6e-15
Identities = 38/104 (36%), Positives = 65/104 (61%)
Frame = +3
Query: 353 TRLKVVLDKCISEFQSAFVPGRSIFDNVMAAIEIIHHMKIKVKGNVGEVALKLDISKAYD 412
+R++ VL+ +SE QSAFV GR IFDN++ + E++ KG +K+D+ KAYD
Sbjct: 309 SRMQGVLNSVVSENQSAFVKGRVIFDNIILSHELVK--SYSRKGISPRCMVKIDLXKAYD 482
Query: 413 RIEWNYLRQVMDKMGFSEQWIKWIMSCVETVDYFVLVNGNVTGP 456
EW +++ +M ++GF +++ W+M+ + T Y NG++T P
Sbjct: 483 SXEWPFIKHLMLELGFPYKFVNWVMAXLTTASYTFNXNGDLTXP 614
>BQ148771
Length = 680
Score = 70.9 bits (172), Expect = 3e-12
Identities = 55/219 (25%), Positives = 100/219 (45%), Gaps = 9/219 (4%)
Frame = -3
Query: 590 SKKATFNFIKDRVWKKINSWSSKCLSKAGREVLIKFVLQSIPTYFMSLFTLPLSLCDEIE 649
+KK+ F+ + +V + +W + LS A R L K V++++P Y M +P + +EI+
Sbjct: 618 TKKSRFS-VYYQVHVMLANWKANHLSLARRVTLAKSVIEAVPLYPMMTTIIPKACIEEIQ 442
Query: 650 KMMNSFWWGHSGSNKKGIHWMSWDKLSMHKKDGGMSFKNLGTFNLAMLGKQGWRILTQSD 709
K+ F WG + +++ H + W+ +S K G+ + L N A + K GW I + S+
Sbjct: 441 KLQRKFVWGDTEVSRR-YHAVGWETMSKPKTIYGLGLRRLDVMNKACIMKLGWSIYSGSN 265
Query: 710 TLIARIYKARYFPRCNFFESELGHNP--SFVWRSICK-----SKFILKAGSRWKIGDGTS 762
+L + + +Y R E P S +W+++ K + ++ + W
Sbjct: 264 SLCTEVMRGKY-QRSESLEEIFLEKPTDSSLWKALVKLWPEIERNLVDSNGNWN------ 106
Query: 763 ISVWN--NYWMRNDVTIFPLDDVATTLANLRVSDCIMPN 799
W W+ DV + + +AT NL I PN
Sbjct: 105 ---WEKLKQWLPFDV-LQRIMAIATPHENLGSDKLIWPN 1
>BE248682 similar to GP|18568269|gb putative gag-pol polyprotein {Zea mays},
partial (1%)
Length = 441
Score = 69.7 bits (169), Expect = 6e-12
Identities = 46/125 (36%), Positives = 63/125 (49%), Gaps = 1/125 (0%)
Frame = +3
Query: 461 RGLRQGDPLSPYLFIICAEGLSALIRKAEARGDINGVKVCNNAPIISHLLFADDCFLFFR 520
RGL+QGDPL+P+LF++ AEG+S L++ A R G V +SHL +ADD
Sbjct: 24 RGLKQGDPLAPFLFLLVAEGISGLMKNAVNRNLFQGFDVKRGGTRVSHLQYADDTLCIGM 203
Query: 521 ANLNKAQKMRNILSVYEKASGQAVNLQKSEFFCSRNVSPADHKNIA-NILGVKTVLGTGK 579
++ ++ +L +E ASG VN KS NV P D A L +
Sbjct: 204 PTVDNLWTLKALLQGFEMASGLKVNFHKSSLI-GINV-PRDFMEAACRFLNCREESIPFI 377
Query: 580 YLGLP 584
YLGLP
Sbjct: 378 YLGLP 392
>TC81772
Length = 982
Score = 58.2 bits (139), Expect(2) = 4e-10
Identities = 19/41 (46%), Positives = 32/41 (77%)
Frame = -2
Query: 731 LGHNPSFVWRSICKSKFILKAGSRWKIGDGTSISVWNNYWM 771
+GHNPS++W S+ + +L+ G WKIGDG+SI++W ++W+
Sbjct: 717 IGHNPSYIWCSV*TLRMVLEEGHHWKIGDGSSINIWTDHWL 595
Score = 25.4 bits (54), Expect(2) = 4e-10
Identities = 10/21 (47%), Positives = 15/21 (70%)
Frame = -1
Query: 686 FKNLGTFNLAMLGKQGWRILT 706
F ++ +L MLGKQ W++LT
Sbjct: 850 FHDIYGLDLVMLGKQVWKLLT 788
>BG646730
Length = 799
Score = 60.1 bits (144), Expect = 5e-09
Identities = 31/109 (28%), Positives = 54/109 (49%)
Frame = +1
Query: 727 FESELGHNPSFVWRSICKSKFILKAGSRWKIGDGTSISVWNNYWMRNDVTIFPLDDVATT 786
F E+G+ PS+ WRS+ K ++ GSRW I +G ++ +W + W+ N +
Sbjct: 373 FALEIGNQPSYAWRSMFNIKDVIDLGSRWSISNGQNVRIWKDDWLPNQTGFKVCSPMVDF 552
Query: 787 LANLRVSDCIMPNQKSWNIPLLNSIFTEQIVEHIVSTPLYPSVREDRLI 835
+S+ I N KS L+ ++F+ + I++ PL + DR I
Sbjct: 553 EEATCISELIDTNTKSSKRDLVRNVFSAFEAKQILNIPLSWRLWPDRRI 699
>BF647389
Length = 404
Score = 43.9 bits (102), Expect = 4e-04
Identities = 40/128 (31%), Positives = 63/128 (48%), Gaps = 4/128 (3%)
Frame = +2
Query: 579 KYLGLPSMIGRSKKATFNFIKDRVWK---KINSWSSKCLSKAGREVLIKFVLQSIPTYFM 635
+YLG+P SKK ++ RV K +I + SSK LS AG LIK VL + Y+
Sbjct: 41 RYLGVPLS---SKKL---YVIQRVKKIICRIEN*SSKLLSYAGSLQLIKIVLFGVQPYWS 202
Query: 636 SLFTLPLSLCDEIEKMMNSFW-WGHSGSNKKGIHWMSWDKLSMHKKDGGMSFKNLGTFNL 694
+F LP + I+ F G SG++K+ + ++ + + + K GG + +L N
Sbjct: 203 QVFVLP*KVIKLIQTTCRIFL*TGKSGTSKRAL--IAREHICLPKTAGGWNVIDLKVXNQ 376
Query: 695 AMLGKQGW 702
+ K W
Sbjct: 377 TAICKLXW 400
>TC80819 weakly similar to GP|10140689|gb|AAG13524.1 putative non-LTR
retroelement reverse transcriptase {Oryza sativa
(japonica cultivar-group)}, partial (2%)
Length = 1262
Score = 37.4 bits (85), Expect(2) = 7e-04
Identities = 16/32 (50%), Positives = 22/32 (68%)
Frame = +1
Query: 689 LGTFNLAMLGKQGWRILTQSDTLIARIYKARY 720
+G FNL++LGK WR+L + L R+ KARY
Sbjct: 40 VGAFNLSLLGKWCWRLLVDKEGLWHRVLKARY 135
Score = 24.6 bits (52), Expect(2) = 7e-04
Identities = 14/49 (28%), Positives = 20/49 (40%), Gaps = 8/49 (16%)
Frame = +3
Query: 730 ELGHNPSFVWRSICKSKFILKAG--------SRWKIGDGTSISVWNNYW 770
E G S W++ICK + + G R +GDG W + W
Sbjct: 159 EGGRQSSMWWKTICKVREGVGEGVGNWFEENIRMVVGDGRDAFFWYDTW 305
>BG587113 weakly similar to PIR|A84888|A8 hypothetical protein At2g45230
[imported] - Arabidopsis thaliana, partial (10%)
Length = 767
Score = 40.0 bits (92), Expect = 0.005
Identities = 32/109 (29%), Positives = 48/109 (43%), Gaps = 7/109 (6%)
Frame = -3
Query: 731 LGHNPSFVWRSICKSKFILKAGSRWKIGDGTSISVWNN--YWMRNDVTIFPLDDV----- 783
LG S+ WRSI ++ ++K G++ IG+G + ++W W VT P
Sbjct: 750 LGSWASYAWRSIHSAQHLIKQGAKVIIGNGENTNIWEREMAWKLTCVTNHPNKHSSRAY* 571
Query: 784 ATTLANLRVSDCIMPNQKSWNIPLLNSIFTEQIVEHIVSTPLYPSVRED 832
A TL P ++ N L+NSIF E I+S + ED
Sbjct: 570 APTLYGYEGCRSDDPMRRERNANLINSIFPEGTRRKILSIHPQGPIGED 424
>BQ149324
Length = 374
Score = 37.0 bits (84), Expect = 0.045
Identities = 23/70 (32%), Positives = 36/70 (50%)
Frame = +2
Query: 922 YISYSNVQVAEIYGVCGARPILLITYSISTKMSRISYSEYFRYCQMMMHLCLVVLYGVYG 981
+I + N QVA ++GVC +L + + RI +S+++ M M L VV GVY
Sbjct: 89 FI*FFNAQVA*MFGVCFHSFLLFLFCYNKIWIVRILFSKHYMIYLMKMQLYFVVCCGVYR 268
Query: 982 SKGIIKSGRK 991
+K K G +
Sbjct: 269 NKETTKCGNE 298
>BF646462
Length = 487
Score = 33.9 bits (76), Expect = 0.38
Identities = 20/41 (48%), Positives = 27/41 (65%)
Frame = -1
Query: 1143 MKTLVSSLYGGKQMRLLIV*LKRPYHQLAPKFW*KYLIVLN 1183
++T SSL G KQMR L +* + P++QLA YL+VLN
Sbjct: 166 LQTXRSSLVGDKQMR*LTL*RELPHYQLALSXIIIYLVVLN 44
>TC77200 similar to GP|22597162|gb|AAN03468.1 bZIP transcription factor ATB2
{Glycine max}, partial (84%)
Length = 1481
Score = 30.0 bits (66), Expect = 5.5
Identities = 17/49 (34%), Positives = 28/49 (56%)
Frame = +2
Query: 216 YVQWLKIILLIFFYRKRVTQQLSLKLSVHPSQWRIMFYLKHLLLLKNLK 264
Y+ ++ II +I FY V LSL ++ S W ++ +L LLL+ +K
Sbjct: 359 YIIYIYIIKIIKFYISAVLIFLSLLFNIFYSVWVLLVFLSLLLLVLIIK 505
>TC77455 similar to GP|22335695|dbj|BAC10549. nine-cis-epoxycarotenoid
dioxygenase1 {Pisum sativum}, partial (43%)
Length = 1865
Score = 29.6 bits (65), Expect = 7.2
Identities = 12/42 (28%), Positives = 23/42 (54%)
Frame = -3
Query: 682 GGMSFKNLGTFNLAMLGKQGWRILTQSDTLIARIYKARYFPR 723
GG+ +++ N+++L K WR+L +L + + Y PR
Sbjct: 951 GGLGVRDIRLVNVSLLAKWWWRLLQDQSSLWKEVLEDIYGPR 826
>TC79696 similar to GP|15216028|emb|CAC51424. amino acid permease AAP3 {Vicia
faba var. minor}, partial (52%)
Length = 1111
Score = 29.3 bits (64), Expect = 9.5
Identities = 11/32 (34%), Positives = 17/32 (52%)
Frame = +1
Query: 1089 VCYLLWIGCICCNWEQWTSRWMQREWLIALIL 1120
+ + LW+ ICC W Q + +R W + IL
Sbjct: 205 ILFALWLHGICCFWRQCARQPARRIWCLKSIL 300
>TC82861 similar to GP|6630450|gb|AAF19538.1| F23N19.10 {Arabidopsis
thaliana}, partial (29%)
Length = 750
Score = 29.3 bits (64), Expect = 9.5
Identities = 12/35 (34%), Positives = 17/35 (48%)
Frame = -1
Query: 640 LPLSLCDEIEKMMNSFWWGHSGSNKKGIHWMSWDK 674
LPL+L FW KKGI+++ WD+
Sbjct: 111 LPLALASSAMDRSTGFWGFCDRKKKKGIYFVDWDR 7
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.342 0.150 0.492
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 40,886,993
Number of Sequences: 36976
Number of extensions: 634451
Number of successful extensions: 5423
Number of sequences better than 10.0: 40
Number of HSP's better than 10.0 without gapping: 3050
Number of HSP's successfully gapped in prelim test: 284
Number of HSP's that attempted gapping in prelim test: 2260
Number of HSP's gapped (non-prelim): 3562
length of query: 1189
length of database: 9,014,727
effective HSP length: 107
effective length of query: 1082
effective length of database: 5,058,295
effective search space: 5473075190
effective search space used: 5473075190
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (22.0 bits)
S2: 64 (29.3 bits)
Medicago: description of AC138131.4


