

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC138130.16 + phase: 0
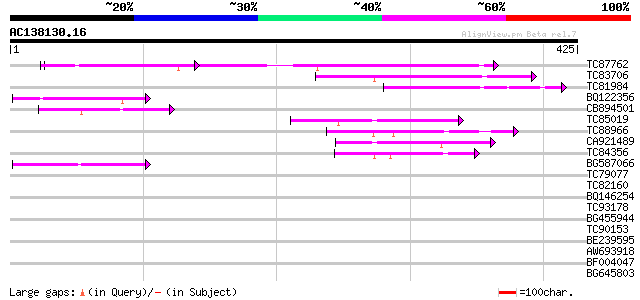
(425 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC87762 similar to GP|21537142|gb|AAM61483.1 unknown {Arabidopsi... 94 1e-19
TC83706 weakly similar to PIR|T48283|T48283 ankyrin-like protein... 82 3e-16
TC81984 homologue to GP|12321945|gb|AAG51002.1 ankyrin-like prot... 71 7e-13
BQ122356 59 3e-09
CB894501 weakly similar to PIR|F86279|F86 hypothetical protein A... 56 3e-08
TC85019 weakly similar to GP|9293890|dbj|BAB01793.1 emb|CAB70981... 55 4e-08
TC88966 similar to GP|21305941|gb|AAM45816.1 NADH dehydrogenase ... 53 2e-07
CA921489 weakly similar to PIR|T47566|T475 hypothetical protein ... 53 3e-07
TC84356 49 4e-06
BG587066 similar to PIR|E84725|E84 ankyrin-like protein [importe... 45 5e-05
TC79077 similar to GP|17529252|gb|AAL38853.1 unknown protein {Ar... 38 0.006
TC82160 37 0.011
BQ146254 weakly similar to GP|11994366|dbj gene_id:MDC16.7~unkno... 36 0.032
TC93178 35 0.042
BG455944 34 0.094
TC90153 similar to GP|20258937|gb|AAM14184.1 unknown protein {Ar... 33 0.16
BE239595 weakly similar to GP|21592507|gb| unknown {Arabidopsis ... 33 0.27
AW693918 similar to GP|9759271|dbj contains similarity to ankyri... 33 0.27
BF004047 weakly similar to GP|15027957|gb| putative arginine/ser... 32 0.61
BG645803 30 1.4
>TC87762 similar to GP|21537142|gb|AAM61483.1 unknown {Arabidopsis
thaliana}, partial (65%)
Length = 1594
Score = 93.6 bits (231), Expect = 1e-19
Identities = 77/345 (22%), Positives = 151/345 (43%), Gaps = 5/345 (1%)
Frame = +1
Query: 27 ETVFHLAVRYGCYDALVFLVQVSNGTNLLHCQDRYGNSVLHLAVSGGRHKMTDFLINKTK 86
+T H AVRYG + L+ G ++ +D+ G + LH+AV G + + ++
Sbjct: 4 KTALHNAVRYGVDRIVKALIVRDPG--IVCIKDKKGQTALHMAVKGQSTSVVEEILQADP 177
Query: 87 LDINTRNSEGMTALDILDQAMDSVESRQLQAIFIRAGGKRSIQSSSFSLELDKNNSPSPA 146
+N R+ +G TAL M + + R ++ + + + N A
Sbjct: 178 TILNERDKKGNTALH-----MATRKGRSQIVSYLLSYAAVDVNAI--------NKQQETA 318
Query: 147 YRLSPSRRYIPNEMEVLTEMVSYDCISPPPVSKSSDSRSPQPQASERFENGTYNPYYVSP 206
L+ Y + +E+ + Y V K ++ + S+
Sbjct: 319 LDLADKLPYGSSALEIQEALSEYGAKYARHVGKVDEAMELKRTVSDI------------- 459
Query: 207 TNLVKQKHHHNKGKIENVNHTKR-----KHYHEMHQEALLNARNTIVLVAVLIATVTFAA 261
KH I+N +R K ++H+EA+ N N++ +VAVL A++ F A
Sbjct: 460 ------KHEVQSQLIQNEKTRRRVSGIAKELKKLHREAVQNTINSVTVVAVLFASIAFLA 621
Query: 262 GISPPGGVYQEGPMRGKSMVGRTSAFKVFAISNNIALFTSLSVVIVLVSIVPFRRKPQTL 321
+ PG +G G+S + F++F + N+ +LF SL+VV+V +++V + + Q
Sbjct: 622 IFNLPGQYIMKGSHIGESNIADHVGFQIFCLLNSTSLFISLAVVVVQITLVAWDTRAQKQ 801
Query: 322 LLIIAHKVMWVAVAFMATGYVAATWVILPHNQGMQWLSVLLLALG 366
++ + +K+MW A A ++A + ++ +W+++ + LG
Sbjct: 802 IVSVVNKLMWAACACTCGAFLAIAFEVVGKK---KWMAITITGLG 927
Score = 49.7 bits (117), Expect = 2e-06
Identities = 30/121 (24%), Positives = 61/121 (49%), Gaps = 2/121 (1%)
Frame = +1
Query: 24 REEETVFHLAVRYGCYDALVFLVQVSNGTNLLHCQDRYGNSVLHLAVSGGRHKMTDFLIN 83
++ +T H+AV+ + ++Q +L+ +D+ GN+ LH+A GR ++ +L++
Sbjct: 97 KKGQTALHMAVKGQSTSVVEEILQAD--PTILNERDKKGNTALHMATRKGRSQIVSYLLS 270
Query: 84 KTKLDINTRNSEGMTALDILDQAMDSVESRQLQAIFIRAGGK--RSIQSSSFSLELDKNN 141
+D+N N + TALD+ D+ + ++Q G K R + ++EL +
Sbjct: 271 YAAVDVNAINKQQETALDLADKLPYGSSALEIQEALSEYGAKYARHVGKVDEAMELKRTV 450
Query: 142 S 142
S
Sbjct: 451 S 453
>TC83706 weakly similar to PIR|T48283|T48283 ankyrin-like protein -
Arabidopsis thaliana, partial (34%)
Length = 746
Score = 82.4 bits (202), Expect = 3e-16
Identities = 53/172 (30%), Positives = 90/172 (51%), Gaps = 6/172 (3%)
Frame = +1
Query: 230 KHYHEMHQEALLNARNTIVLVAVLIATVTFAAGISPPGGVYQE------GPMRGKSMVGR 283
K ++M E L NA N+ +VAVLIATV FAA + PG Q G G++ +
Sbjct: 55 KRINKMQAEGLNNAINSNTVVAVLIATVAFAAIFTVPGQYPQNTKNLAPGMSPGEANIAP 234
Query: 284 TSAFKVFAISNNIALFTSLSVVIVLVSIVPFRRKPQTLLLIIAHKVMWVAVAFMATGYVA 343
F +F I ++ ALF SL+VVIV S+V R+ + + + +K+MW+A ++ ++A
Sbjct: 235 NIEFLIFVIFDSTALFISLAVVIVQTSVVVIEREAKKQMTAVINKLMWIACVLISVAFLA 414
Query: 344 ATWVILPHNQGMQWLSVLLLALGGGSLGTIFIGLSVMLVEHSLRKSKWRKKR 395
+++++ + L++ ALG + L ++ H L S+ R +R
Sbjct: 415 MSYIVVGDQKE---LAIAATALGTVIMAATLGTLCYWVIAHRLEASRLRSQR 561
>TC81984 homologue to GP|12321945|gb|AAG51002.1 ankyrin-like protein;
93648-91299 {Arabidopsis thaliana}, partial (22%)
Length = 649
Score = 71.2 bits (173), Expect = 7e-13
Identities = 44/138 (31%), Positives = 78/138 (55%), Gaps = 1/138 (0%)
Frame = +3
Query: 281 VGRTSAFKVFAISNNIALFTSLSVVIVLVSIVPFRRKPQTLLLIIAHKVMWVAVAFMATG 340
V SAFK+F I N IALFTSL+VV+V +++V K + ++ + +K+MW+A +
Sbjct: 3 VASYSAFKIFFIFNAIALFTSLAVVVVQITLVRGETKAEKRVVEVINKLMWLASVCTSVA 182
Query: 341 YVAATWVILPHNQGMQWLSVLLLALGGGSLGTIFIGLSVMLVEHSLRKSKWRKKRKEIGD 400
++A++++++ +W ++L+ +GG + + IG V S R RKK K++
Sbjct: 183 FIASSYIVVGRRN--EWAAILVTVVGGVIISGV-IGTMTYYVVRSKRTRSMRKKEKQL-- 347
Query: 401 GAAESDKES-ENSDFQSS 417
A S S +S+F +S
Sbjct: 348 -ARRSGSNSWHHSEFSNS 398
>BQ122356
Length = 447
Score = 59.3 bits (142), Expect = 3e-09
Identities = 36/106 (33%), Positives = 63/106 (58%), Gaps = 3/106 (2%)
Frame = +3
Query: 3 GKVSILDDFVSGSAASFHYLTREEETVFHLAVRYGCYDALVFLVQVSNGTNLL-HCQDRY 61
G V ++++ + S + ++ ++ HL VRY +AL +LVQ NG L H QD+
Sbjct: 90 GHVEVVEELKNAKPCSIQKIG-DDGSLLHLCVRYNHLEALKYLVQSVNGAQEL*HVQDKE 266
Query: 62 GNSVLHLAVSGGRHKMTDFLIN--KTKLDINTRNSEGMTALDILDQ 105
GN++LH AV+ + + FL++ + + NT N++G+T LDIL++
Sbjct: 267 GNTILH*AVNLKQIQTIKFLLSLPQMRTAANTLNNKGLTPLDILNK 404
>CB894501 weakly similar to PIR|F86279|F86 hypothetical protein AAF43949.1
[imported] - Arabidopsis thaliana, partial (26%)
Length = 812
Score = 55.8 bits (133), Expect = 3e-08
Identities = 34/112 (30%), Positives = 59/112 (52%), Gaps = 10/112 (8%)
Frame = +3
Query: 22 LTREEETVFHLAVRYGCYDALVFLVQVSNGT----------NLLHCQDRYGNSVLHLAVS 71
+T ET H+AV+Y + L L+ T +L+ +D GN++LH++V
Sbjct: 336 VTVRSETALHIAVKYKQFHVLEILLGWLRRTCHRRSHHKEKRVLNWEDEAGNTILHMSVL 515
Query: 72 GGRHKMTDFLINKTKLDINTRNSEGMTALDILDQAMDSVESRQLQAIFIRAG 123
+ LI+ + +DIN +N + TALDI++Q V S +++ + I+AG
Sbjct: 516 NSFPQAVGLLID-SNIDINAKNLDEQTALDIVEQIQSQVYSAEMKDMLIKAG 668
Score = 30.0 bits (66), Expect = 1.8
Identities = 15/47 (31%), Positives = 24/47 (50%)
Frame = +3
Query: 59 DRYGNSVLHLAVSGGRHKMTDFLINKTKLDINTRNSEGMTALDILDQ 105
+ YG S +HLA+ H+M ++ K + + EG+T L I Q
Sbjct: 135 NEYGLSPIHLALQNKYHRMVCRFVDINKDLVRVKGREGLTPLHIATQ 275
>TC85019 weakly similar to GP|9293890|dbj|BAB01793.1
emb|CAB70981.1~gene_id:MVE11.3~similar to unknown
protein {Arabidopsis thaliana}, partial (10%)
Length = 663
Score = 55.5 bits (132), Expect = 4e-08
Identities = 40/138 (28%), Positives = 71/138 (50%), Gaps = 8/138 (5%)
Frame = +2
Query: 211 KQKHHHNKGKIENVNHTKRKHYHEMHQEALLNARN-------TIVLVAVLIATVTFAAGI 263
K H +K N T + + E H+ L +N + +LVA LIAT+TFAA I
Sbjct: 221 KLDHPLHKDVKNNDGKTAWQVFKEEHKTLLEEGKNWMKDTSNSCMLVATLIATITFAAAI 400
Query: 264 SPPGGVYQEGPMRGKSMVGRTSAFKVFAISNNIALFTSLSVVIVLVSIVPFRRKPQTLLL 323
+ PGG Q+ +G + F +F +S+ +ALF+S+ +++ +SI+ R + ++
Sbjct: 401 TVPGGNNQD---KGIPIFLSDKTFMLFIVSDALALFSSMVSLLMFLSIIHGRYAKEDFVV 571
Query: 324 IIAHK-VMWVAVAFMATG 340
+ + ++ +A F A G
Sbjct: 572 ALPKRLILGMAALFFAVG 625
>TC88966 similar to GP|21305941|gb|AAM45816.1 NADH dehydrogenase subunit 2
{Neoheterandria umbratilis}, partial (4%)
Length = 915
Score = 53.1 bits (126), Expect = 2e-07
Identities = 46/165 (27%), Positives = 79/165 (47%), Gaps = 21/165 (12%)
Frame = -2
Query: 238 EALLNARNTIVLVAVLIATVTFAAGISPPGGVYQ----EGPMRGKSMVGRTSA------- 286
E L + +++I L A LIAT+TF+ +PPGGV Q + GK ++ +
Sbjct: 821 EWLKDMKSSISLTASLIATLTFSLATNPPGGVVQASVGDSNECGKILISTINTTICVGEA 642
Query: 287 ---------FKVFAISNNIALFTSLSVVIVLVSIVPFRRKPQTLLLIIAHKVMWVAVAFM 337
+ F I N I SLSV++VLVS +P K LL + M + ++ +
Sbjct: 641 ILATRSHDKYLAFLICNTICFIASLSVILVLVSGIPIDNKFSMWLLSMG---MSIILSSL 471
Query: 338 ATGYVAATWVILPHNQGMQWLSVLLLALGGGSLG-TIFIGLSVML 381
A Y+ A ++ P++ + +L G G ++F+ ++V+L
Sbjct: 470 ALTYLFAANMVTPNS--------IWNSLSGNGFGISLFVWIAVVL 360
>CA921489 weakly similar to PIR|T47566|T475 hypothetical protein F24B22.30 -
Arabidopsis thaliana, partial (10%)
Length = 809
Score = 52.8 bits (125), Expect = 3e-07
Identities = 37/126 (29%), Positives = 67/126 (52%), Gaps = 6/126 (4%)
Frame = -3
Query: 245 NTIVLVAVLIATVTFAAGISPPGGVYQEGPMRGKSMVGRTSAFKVFAISNNIALFTSLSV 304
N+ +LVA LIAT+ FAA I+ PGG Q+ +G + + F VF +S+ +ALF+S++
Sbjct: 558 NSCMLVATLIATIAFAAAITVPGGNNQD---KGIPIFLSDNTFMVFVVSDALALFSSMAS 388
Query: 305 VIVLVSIVPFRRKPQTLL------LIIAHKVMWVAVAFMATGYVAATWVILPHNQGMQWL 358
+++ ++I+ R + + LI+ ++ AV + AA ++L +
Sbjct: 387 LLMFLAILNARYTEEDFMMALPERLILGMASLFFAVVTTMVAFGAALSMLLKERLTWAPI 208
Query: 359 SVLLLA 364
+ LLA
Sbjct: 207 PIALLA 190
>TC84356
Length = 670
Score = 48.9 bits (115), Expect = 4e-06
Identities = 40/130 (30%), Positives = 58/130 (43%), Gaps = 21/130 (16%)
Frame = +3
Query: 244 RNTIVLVAVLIATVTFAAGISPPGGVYQE--------GPMRGKSMVGRT----------- 284
+ +I L A LIAT+TF+ +PPGGV Q + MV T
Sbjct: 114 KGSISLTASLIATLTFSLATNPPGGVVQASLDDSNYCSTILNTRMVNTTICLGEAILATR 293
Query: 285 --SAFKVFAISNNIALFTSLSVVIVLVSIVPFRRKPQTLLLIIAHKVMWVAVAFMATGYV 342
+ F I N SLSV+ VLVS +P K LL I VM + ++ +A Y+
Sbjct: 294 LKDDYLAFLICNTFCFIASLSVIFVLVSGIPINNKFLIWLLSI---VMSIILSGLALTYL 464
Query: 343 AATWVILPHN 352
A ++ P++
Sbjct: 465 FAAGMVTPNS 494
>BG587066 similar to PIR|E84725|E84 ankyrin-like protein [imported] -
Arabidopsis thaliana, partial (32%)
Length = 656
Score = 45.1 bits (105), Expect = 5e-05
Identities = 26/103 (25%), Positives = 54/103 (52%)
Frame = +2
Query: 3 GKVSILDDFVSGSAASFHYLTREEETVFHLAVRYGCYDALVFLVQVSNGTNLLHCQDRYG 62
G + ++ +S ++ ++ +T H+AV+ + L+ LV+ +L+ +D G
Sbjct: 161 GHLEVVKALLSKDPSTGFRTDKKGQTALHMAVKGQNEEILLELVKPDPA--VLNLEDNKG 334
Query: 63 NSVLHLAVSGGRHKMTDFLINKTKLDINTRNSEGMTALDILDQ 105
N+ LH+A GR++ L++ ++IN N G T LD+ ++
Sbjct: 335 NTALHIAAKKGRNQNVRRLLSTEGININATNKAGETPLDVAEK 463
>TC79077 similar to GP|17529252|gb|AAL38853.1 unknown protein {Arabidopsis
thaliana}, partial (38%)
Length = 2200
Score = 38.1 bits (87), Expect = 0.006
Identities = 36/135 (26%), Positives = 57/135 (41%), Gaps = 13/135 (9%)
Frame = +3
Query: 3 GKVSILDDFVSGSAASFHYLTREEETVFHLAVR------YGCYDALVFLVQVSNGTNLLH 56
G++S ++ +S + ET H AV + D V L++ TN H
Sbjct: 1410 GQLSAVNALISLFPTLISHRNNAGETFLHKAVSGFQTHAFRRLDRQVELLKKLLSTNHFH 1589
Query: 57 CQD------RYGNSVLHLAVSGGRH-KMTDFLINKTKLDINTRNSEGMTALDILDQAMDS 109
++ G + LH+A+ G H + L+ +++N + GMT LD L Q +S
Sbjct: 1590 VEEIINIKNNDGRTALHMAIIGNIHIDLVQLLMTAPFINLNICDVHGMTPLDYLKQNPNS 1769
Query: 110 VESRQLQAIFIRAGG 124
S L I AGG
Sbjct: 1770 SNSDILIKKLISAGG 1814
Score = 38.1 bits (87), Expect = 0.006
Identities = 26/99 (26%), Positives = 48/99 (48%), Gaps = 1/99 (1%)
Frame = +3
Query: 3 GKVSILDDFVSGSAASF-HYLTREEETVFHLAVRYGCYDALVFLVQVSNGTNLLHCQDRY 61
G + IL++ ++ + Y E T H A G + + +L ++ ++++ D
Sbjct: 1206 GNLKILEELLANCSDDILAYRDAEGSTALHAAAARGKVEVVKYL---ASSFDIINSTDHQ 1376
Query: 62 GNSVLHLAVSGGRHKMTDFLINKTKLDINTRNSEGMTAL 100
GN+ LH+A S G+ + LI+ I+ RN+ G T L
Sbjct: 1377 GNTALHVAASRGQLSAVNALISLFPTLISHRNNAGETFL 1493
>TC82160
Length = 839
Score = 37.4 bits (85), Expect = 0.011
Identities = 22/63 (34%), Positives = 33/63 (51%)
Frame = +3
Query: 290 FAISNNIALFTSLSVVIVLVSIVPFRRKPQTLLLIIAHKVMWVAVAFMATGYVAATWVIL 349
F I N I SLSV+++LVS +P K LL I M + + +A Y+ A ++
Sbjct: 246 FLICNTICFIASLSVILLLVSGIPINNKFSMWLLSIG---MSIVITSLALTYLFAAKMVN 416
Query: 350 PHN 352
PH+
Sbjct: 417 PHS 425
>BQ146254 weakly similar to GP|11994366|dbj gene_id:MDC16.7~unknown protein
{Arabidopsis thaliana}, partial (19%)
Length = 408
Score = 35.8 bits (81), Expect = 0.032
Identities = 18/44 (40%), Positives = 26/44 (58%)
Frame = +3
Query: 226 HTKRKHYHEMHQEALLNARNTIVLVAVLIATVTFAAGISPPGGV 269
HTK ++ + L R +VA+ IAT+TF G++PPGGV
Sbjct: 147 HTKTRN------DWLEKMRGNFSVVAIFIATITFQMGLNPPGGV 260
>TC93178
Length = 661
Score = 35.4 bits (80), Expect = 0.042
Identities = 29/98 (29%), Positives = 43/98 (43%), Gaps = 31/98 (31%)
Frame = +3
Query: 244 RNTIVLVAVLIATVTFAAGISPPGGV-----------------------------YQEGP 274
R + L+A +IAT+TF ++PPGGV Y E
Sbjct: 366 RGNLGLIATVIATMTFQMVLNPPGGVMSIKDGTDPPSTNANPPDADDSDKNCTFIYGERL 545
Query: 275 MRGKSM--VGRTSAFKVFAISNNIALFTSLSVVIVLVS 310
G+++ VG + + F ISN I SLS+ ++LVS
Sbjct: 546 CPGEAVLAVGDSYGYFQFLISNTIWFVASLSICLLLVS 659
>BG455944
Length = 654
Score = 34.3 bits (77), Expect = 0.094
Identities = 14/26 (53%), Positives = 19/26 (72%)
Frame = +1
Query: 244 RNTIVLVAVLIATVTFAAGISPPGGV 269
R + +VA+ IAT+TF I+PPGGV
Sbjct: 220 RGNMSIVAIFIATITFQMAINPPGGV 297
>TC90153 similar to GP|20258937|gb|AAM14184.1 unknown protein {Arabidopsis
thaliana}, partial (70%)
Length = 721
Score = 33.5 bits (75), Expect = 0.16
Identities = 22/65 (33%), Positives = 33/65 (49%), Gaps = 4/65 (6%)
Frame = +3
Query: 181 SDSRSPQPQASERFENGTYNP----YYVSPTNLVKQKHHHNKGKIENVNHTKRKHYHEMH 236
S + SP +S F + T P YY PT+L+K +HH + + NH H+H +H
Sbjct: 27 SHTSSPTHPSSIIFPSPTLPPFPPSYYARPTSLLK-RHHSTRPSHHHNNH----HHHHVH 191
Query: 237 QEALL 241
+ LL
Sbjct: 192 EFQLL 206
>BE239595 weakly similar to GP|21592507|gb| unknown {Arabidopsis thaliana},
partial (44%)
Length = 651
Score = 32.7 bits (73), Expect = 0.27
Identities = 29/105 (27%), Positives = 44/105 (41%), Gaps = 6/105 (5%)
Frame = +1
Query: 284 TSAFKVFAISNNIALFTSLSVVIVLVSIVPFRRKPQTLLLIIAHKVMWVAVAFMATGYVA 343
T AF F ++N I +L ++ + F K L LI M +A+A G
Sbjct: 199 TPAFVYFVVANGIVSLHNLEMIAKHILGPKFHNKGLQLALIAVFDTMALALASSGDGAAT 378
Query: 344 ATWVILPH-NQGMQWLSVL-----LLALGGGSLGTIFIGLSVMLV 382
A + + N +W + GGGSL FIGL ++L+
Sbjct: 379 AMSELGRNGNSHAKWNKICDKFESYCNRGGGSLIASFIGLILLLI 513
>AW693918 similar to GP|9759271|dbj contains similarity to
ankyrin~gene_id:MRI1.10 {Arabidopsis thaliana}, partial
(21%)
Length = 375
Score = 32.7 bits (73), Expect = 0.27
Identities = 16/38 (42%), Positives = 25/38 (65%)
Frame = +3
Query: 63 NSVLHLAVSGGRHKMTDFLINKTKLDINTRNSEGMTAL 100
NS LH + + G H++ +L+ ++ +DIN RN G TAL
Sbjct: 87 NSPLHYSAAHGHHEIV-YLLLESGVDINLRNYRGQTAL 197
>BF004047 weakly similar to GP|15027957|gb| putative arginine/serine-rich
protein {Arabidopsis thaliana}, partial (11%)
Length = 835
Score = 31.6 bits (70), Expect = 0.61
Identities = 32/89 (35%), Positives = 42/89 (46%), Gaps = 2/89 (2%)
Frame = -3
Query: 122 AGGKRSIQSS-SFSLELDKNNSPSPAYRLSPSRRYIPNEMEVLTEMVSYDCISPP-PVSK 179
+ +RS+ +S SFS ++SPS SPS R P ++ SPP P S
Sbjct: 290 SSSERSVSTSTSFSCSSSLSSSPST----SPSSRSPP-------PLIFPPSGSPPCPPST 144
Query: 180 SSDSRSPQPQASERFENGTYNPYYVSPTN 208
SS SRSP P S GT P SP++
Sbjct: 143 SSSSRSPPPPTSTSPTTGT-PPRSTSPSS 60
>BG645803
Length = 652
Score = 30.4 bits (67), Expect = 1.4
Identities = 21/72 (29%), Positives = 33/72 (45%), Gaps = 1/72 (1%)
Frame = +2
Query: 24 REEETVFHLAVRYGC-YDALVFLVQVSNGTNLLHCQDRYGNSVLHLAVSGGRHKMTDFLI 82
R +T LA Y D + +V G LL + G +H+A G KMT FL
Sbjct: 320 RYGDTALALAACYNAKIDIVKCMVDSKMGQMLLMKHNTNGELPVHMAAGKGHKKMTSFLY 499
Query: 83 NKTKLDINTRNS 94
++T ++ ++S
Sbjct: 500 SETPGEVFKKDS 535
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.318 0.133 0.383
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 13,292,363
Number of Sequences: 36976
Number of extensions: 204263
Number of successful extensions: 1545
Number of sequences better than 10.0: 60
Number of HSP's better than 10.0 without gapping: 1491
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1532
length of query: 425
length of database: 9,014,727
effective HSP length: 99
effective length of query: 326
effective length of database: 5,354,103
effective search space: 1745437578
effective search space used: 1745437578
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 60 (27.7 bits)
Medicago: description of AC138130.16


