

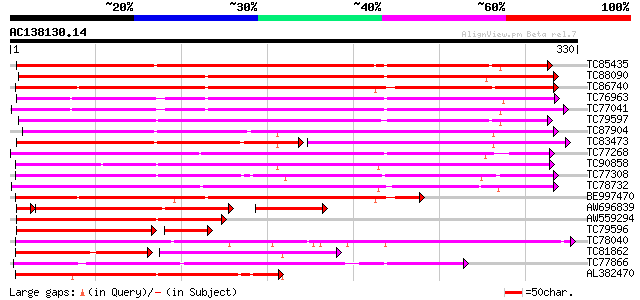
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC138130.14 + phase: 0
(330 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC85435 similar to PIR|T16995|T16995 probable cinnamyl-alcohol d... 254 4e-68
TC88090 similar to PIR|F96709|F96709 probable reductase T26J14.1... 253 9e-68
TC86740 weakly similar to PIR|D84747|D84747 probable cinnamoyl-C... 238 2e-63
TC76963 GP|3716262|emb|CAA03625.1 unnamed protein product {unide... 233 6e-62
TC77041 similar to GP|3716262|emb|CAA03625.1 unnamed protein pro... 230 7e-61
TC79597 weakly similar to PIR|C96552|C96552 hypothetical protein... 224 5e-59
TC87904 similar to GP|5852933|gb|AAD54273.1| dihydroflavonol-4-r... 222 2e-58
TC83473 similar to GP|5852933|gb|AAD54273.1| dihydroflavonol-4-r... 150 7e-57
TC77268 weakly similar to GP|10177026|dbj|BAB10264. dihydroflavo... 214 3e-56
TC90858 anthocyanidin reductase [Medicago truncatula] 209 1e-54
TC77308 PIR|S66262|S66262 vestitone reductase - alfalfa, complete 198 3e-51
TC78732 weakly similar to PIR|C84436|C84436 probable cinnamoyl-C... 193 7e-50
BE997470 similar to PIR|D84747|D84 probable cinnamoyl-CoA reduct... 182 2e-46
AW696839 similar to PIR|T16995|T16 probable cinnamyl-alcohol deh... 133 8e-43
AW559294 similar to GP|21594240|gb cinnamyl-alcohol dehydrogenas... 149 1e-36
TC79596 similar to GP|21594240|gb|AAM65984.1 cinnamyl-alcohol de... 108 3e-30
TC78040 similar to GP|23495824|dbj|BAC20034. putative NADPH HC t... 127 8e-30
TC81862 weakly similar to PIR|T05749|T05749 hypothetical protein... 77 2e-29
TC77866 similar to GP|18266655|gb|AAL67601.1 putative cinnamoyl-... 118 3e-27
AL382470 similar to PIR|S66262|S66 vestitone reductase - alfalfa... 114 7e-26
>TC85435 similar to PIR|T16995|T16995 probable cinnamyl-alcohol
dehydrogenase (EC 1.1.1.195) - apple tree, complete
Length = 1260
Score = 254 bits (648), Expect = 4e-68
Identities = 137/316 (43%), Positives = 204/316 (64%), Gaps = 4/316 (1%)
Frame = +3
Query: 5 VCVTGASGFLASWLIKRLLLSGYHVIGTVRDLGKKQKVEHLWKLEGATERLKLVQADLME 64
VCVTGASG++ASWL++ LL GY V TVRD +KV+HL KL+GA ERL+L +A+L+E
Sbjct: 111 VCVTGASGYIASWLVRLLLHRGYTVKATVRDPNDPKKVDHLVKLDGAKERLQLFKANLLE 290
Query: 65 ENSFDNAIMGCKGVFHIASPVLNHISNDPKAEILEPAVQGTLNVLRSCRKNPALVRVVLA 124
E +FD+ + GC GVFH ASP H DP+AE+++PA++GTLNVL SC K+P+L RVVL
Sbjct: 291 EGAFDSVVQGCHGVFHTASP-FYHDVKDPQAELIDPALKGTLNVLNSCAKSPSLKRVVLT 467
Query: 125 SSSSAVRVRA-DFDPNIPIDESSWSSLELCEKLQAWYPMSKTLAEKAAWDYCKENGIDLV 183
SS +AV P++ +DE+ ++ + C K WY +SKTLAE+AAW + KEN ID+V
Sbjct: 468 SSIAAVAYNGKPRTPDVVVDETWFTDADFCAKSNLWYVVSKTLAEEAAWKFVKENNIDMV 647
Query: 184 TILPSFIIGPNLPTDLCSTASDVLGLFKGETEKFQWHGRMGYVHIDDVALCHILLYENKA 243
TI P+ +IGP L L ++A+ +L L G + F + G+V++ DVA HIL YEN +
Sbjct: 648 TINPAMVIGPLLQPVLNTSAAAILNLING-AQTFP-NASFGWVNVKDVANAHILAYENAS 821
Query: 244 SDGRYLCSSKIMDNDDLVGMLANRYPGFPIPKRFKKLDRPH---YELNTGKLESLGFKFK 300
+ GR+ ++ ++V +L YP +P++ D+P+ Y+++ K +SLG ++
Sbjct: 822 ASGRHCLVERVAHYSEVVRILRELYPSLQLPEKCAD-DKPYVPIYQVSKEKAKSLGLEYT 998
Query: 301 SVEEMFDDCFASFVEQ 316
+E + S E+
Sbjct: 999 PLEVSIKETVESLKEK 1046
>TC88090 similar to PIR|F96709|F96709 probable reductase T26J14.11
[imported] - Arabidopsis thaliana, complete
Length = 1284
Score = 253 bits (645), Expect = 9e-68
Identities = 135/317 (42%), Positives = 198/317 (61%), Gaps = 3/317 (0%)
Frame = +1
Query: 6 CVTGASGFLASWLIKRLLLSGYHVIGTVRDLGKKQKVEHLWKLEGATERLKLVQADLMEE 65
CVTG +GF+A++L+K LL G+ V TVR+ +KV +L +L G ERLK+++ADLM E
Sbjct: 97 CVTGGTGFIAAYLVKALLEKGHIVRTTVRNPDDLEKVGYLTQLSGDKERLKILKADLMVE 276
Query: 66 NSFDNAIMGCKGVFHIASPVLNHISNDPKAEILEPAVQGTLNVLRSCRKNPALVRVVLAS 125
SFD A+ G GVFH ASPV+ N+ +A +++P ++GT NVL SC K + RVVL S
Sbjct: 277 GSFDEAVTGVDGVFHTASPVIVPYDNNIQATLIDPCIKGTQNVLNSCIK-ANVKRVVLTS 453
Query: 126 SSSAVRVRADFDPNIPIDESSWSSLELCEKLQAWYPMSKTLAEKAAWDYCKENGIDLVTI 185
S S++R R D P++ES WS E C++ WY +KTL E+ AW +E+G+DLV +
Sbjct: 454 SCSSIRYRDDVQQVSPLNESHWSDPEYCKRYNLWYAYAKTLGEREAWRIAEESGLDLVVV 633
Query: 186 LPSFIIGPNLPTDLCSTASDVLGLFKGETEKFQWHGRMGYVHIDDVALCHILLYENKASD 245
PSF++GP L ST +L + KG ++ + +G+VHIDDV HIL E +
Sbjct: 634 NPSFVVGPLLAPQPASTLLMILSIVKGSRGEYP-NTTVGFVHIDDVIAAHILAMEEPKAS 810
Query: 246 GRYLCSSKIMDNDDLVGMLANRYPGFPIPKR--FKKLDRPHYELNTGKLESLGF-KFKSV 302
GR +CSS + ++ ML +YP +P + ++ D + ++T K+ LGF +FKS+
Sbjct: 811 GRLVCSSTVAHWSQIIQMLQAKYPSYPYETKCSSQEGDNNTHSMDTTKITQLGFSQFKSL 990
Query: 303 EEMFDDCFASFVEQGHL 319
E+MFDDC SF ++G L
Sbjct: 991 EQMFDDCIKSFQDKGFL 1041
>TC86740 weakly similar to PIR|D84747|D84747 probable cinnamoyl-CoA
reductase [imported] - Arabidopsis thaliana, partial
(85%)
Length = 1198
Score = 238 bits (608), Expect = 2e-63
Identities = 133/320 (41%), Positives = 198/320 (61%), Gaps = 4/320 (1%)
Frame = +1
Query: 4 KVCVTGASGFLASWLIKRLLLSGYHVIGTVRDLGKKQKVEHLWKLEGATERLKLVQADLM 63
KVCVTGA GF+ASWL+K LL GY V GTVR+ G K EHL KLE A+E L L +AD++
Sbjct: 88 KVCVTGAGGFVASWLVKLLLSKGYFVHGTVREPGSP-KYEHLLKLEKASENLTLFKADIL 264
Query: 64 EENSFDNAIMGCKGVFHIASPVLNHISNDPKAEILEPAVQGTLNVLRSCRKNPALVRVVL 123
+ S +AI+GC VFH+ASPV + + +P+ E++EPAV+GT NVL +C K + RVV
Sbjct: 265 DYESVYSAIVGCSAVFHVASPVPSTVVPNPEVEVIEPAVKGTANVLEACLK-ANVERVVF 441
Query: 124 ASSSSAVRVRADFDPNIPIDESSWSSLELCEKLQAWYPMSKTLAEKAAWDYCKENGIDLV 183
SS++AV + + + IDES WS + C+ Q WY +KT AE+ A+++ K G+++V
Sbjct: 442 VSSAAAVAINPNLPKDKAIDESCWSDKDYCKNTQNWYCYAKTEAEEQAFNFAKRTGLNVV 621
Query: 184 TILPSFIIGPNLPTDLCSTASDVLGLFK----GETEKFQWHGRMGYVHIDDVALCHILLY 239
TI P+ ++GP L + S++ ++ + K K +W V + DV +L Y
Sbjct: 622 TICPTLVLGPILQSTTNSSSLALIKILKEGHDSLENKLRW-----IVDVRDVVNAILLAY 786
Query: 240 ENKASDGRYLCSSKIMDNDDLVGMLANRYPGFPIPKRFKKLDRPHYELNTGKLESLGFKF 299
EN +DGRY+C+S ++ DLV L + YP + P + ++D + L++ KL+SLG+KF
Sbjct: 787 ENHKADGRYICTSHTINTRDLVERLKSIYPNYKYPTNYIEMD-DYKMLSSEKLQSLGWKF 963
Query: 300 KSVEEMFDDCFASFVEQGHL 319
+ +EE D S+ E G L
Sbjct: 964 RPLEETLIDSVESYKEAGLL 1023
>TC76963 GP|3716262|emb|CAA03625.1 unnamed protein product {unidentified},
complete
Length = 1468
Score = 233 bits (595), Expect = 6e-62
Identities = 131/319 (41%), Positives = 184/319 (57%), Gaps = 3/319 (0%)
Frame = +1
Query: 5 VCVTGASGFLASWLIKRLLLSGYHVIGTVRDLGKKQKVEHLWKLEGATERLKLVQADLME 64
+CVTGA GF+ASW++K LL GY V GT+R+ K HL KLEGA ERL LV+ DL++
Sbjct: 196 ICVTGAGGFIASWMVKLLLEKGYTVRGTLRN-PDDPKNGHLKKLEGAKERLTLVKVDLLD 372
Query: 65 ENSFDNAIMGCKGVFHIASPVLNHISNDPKAEILEPAVQGTLNVLRSCRKNPALVRVVLA 124
NS A+ GC GVFH ASPV D E++EPAV G NV+ + + + RVV
Sbjct: 373 LNSVKEAVNGCHGVFHTASPV-----TDNPEEMVEPAVNGAKNVIIAGAE-AKVRRVVFT 534
Query: 125 SSSSAVRVRADFDPNIPIDESSWSSLELCEKLQAWYPMSKTLAEKAAWDYCKENGIDLVT 184
SS AV + + ++ +DES WS LE C+K + WY K +AE AAWD KE G+DLV
Sbjct: 535 SSIGAVYMDPNRSVDVEVDESCWSDLEFCKKTKNWYCYGKAVAEAAAWDVAKEKGVDLVV 714
Query: 185 ILPSFIIGPNLPTDLCSTASDVLGLFKGETEKFQWHGRMGYVHIDDVALCHILLYENKAS 244
+ P ++GP L + ++ +L G + + + YVH+ DVAL HIL+YE ++
Sbjct: 715 VNPVLVLGPLLQPTINASTIHILKYLTGSAKTYA-NATQAYVHVRDVALAHILVYEKPSA 891
Query: 245 DGRYLCSSKIMDNDDLVGMLANRYPGFPIPKRFKKLDRPHYE---LNTGKLESLGFKFKS 301
GRYLC+ + +LV +LA +P +PIP + P + + KL+ LG +F
Sbjct: 892 SGRYLCAETSLHRGELVEILAKYFPEYPIPTKCSDEKNPRVKPHIFSNKKLKDLGLEFTP 1071
Query: 302 VEEMFDDCFASFVEQGHLT 320
V E + S +QGHL+
Sbjct: 1072VSECLYETVKSLQDQGHLS 1128
>TC77041 similar to GP|3716262|emb|CAA03625.1 unnamed protein product
{unidentified}, partial (91%)
Length = 1447
Score = 230 bits (586), Expect = 7e-61
Identities = 129/327 (39%), Positives = 187/327 (56%), Gaps = 3/327 (0%)
Frame = +2
Query: 2 EHKVCVTGASGFLASWLIKRLLLSGYHVIGTVRDLGKKQKVEHLWKLEGATERLKLVQAD 61
+ VCVTGA GF+ASWL+K LL GY V GTVR+ + K HL +LEGA ERL L + D
Sbjct: 197 DQTVCVTGAGGFIASWLVKLLLERGYTVRGTVRN-PEDPKNGHLKELEGARERLTLHKVD 373
Query: 62 LMEENSFDNAIMGCKGVFHIASPVLNHISNDPKAEILEPAVQGTLNVLRSCRKNPALVRV 121
L++ S + + GC GVFH ASPV D E+LEPAV GT NV+ + + + RV
Sbjct: 374 LLDLQSIQSVVHGCHGVFHTASPV-----TDNPDEMLEPAVNGTKNVIIASAE-AKVRRV 535
Query: 122 VLASSSSAVRVRADFDPNIPIDESSWSSLELCEKLQAWYPMSKTLAEKAAWDYCKENGID 181
V SS V + + ++ +DES WS LE C+ + WY KT+AE++AWD KEN +D
Sbjct: 536 VFTSSIGTVYMDPNTSRDVVVDESYWSDLEHCKNTKNWYCYGKTVAEQSAWDIAKENQVD 715
Query: 182 LVTILPSFIIGPNLPTDLCSTASDVLGLFKGETEKFQWHGRMGYVHIDDVALCHILLYEN 241
LV + P ++GP L + ++ +L G + + + YVH+ DVAL H+L+YE
Sbjct: 716 LVVVNPVVVLGPLLQPTINASTIHILKYLNGAAKTYV-NATQSYVHVKDVALAHLLVYET 892
Query: 242 KASDGRYLCSSKIMDNDDLVGMLANRYPGFPIPKRFKKLDRPH---YELNTGKLESLGFK 298
++ GRY+C + ++V +LA +P +P+P + P Y+ + KL+ LG +
Sbjct: 893 NSASGRYICCETALHRGEVVEILAKYFPEYPLPTKCSDEKNPRVKPYKFSNQKLKDLGLE 1072
Query: 299 FKSVEEMFDDCFASFVEQGHLTTLPHQ 325
F V++ D S E+GHL P Q
Sbjct: 1073FTPVKQCLYDTVRSLQEKGHLPIPPMQ 1153
>TC79597 weakly similar to PIR|C96552|C96552 hypothetical protein F5D21.12
[imported] - Arabidopsis thaliana, partial (36%)
Length = 1055
Score = 224 bits (570), Expect = 5e-59
Identities = 129/316 (40%), Positives = 192/316 (59%), Gaps = 5/316 (1%)
Frame = +1
Query: 6 CVTGASGFLASWLIKRLLLSGYHVIGTVRDLGKKQKVEHLW-KLEGATERLKLVQADLME 64
C TGA+GF+ASW++K LL GY V T K W L+GA ERL+L +ADL+E
Sbjct: 1 CRTGANGFIASWIVKFLLQCGYTVRATCSSPK*F*KG*SPWLNLDGAKERLQLFKADLLE 180
Query: 65 ENSFDNAIMGCKGVFHIASPVLNHISNDPKAEILEPAVQGTLNVLRSCRKNPALVRVVLA 124
E SFD+ + GC GVFH ASPV + NDP+AE+++PA++GTLNVL+SC K+P++ RV+L
Sbjct: 181 EGSFDSVVEGCDGVFHTASPV-RFVVNDPQAELIDPALKGTLNVLQSCAKSPSVKRVILT 357
Query: 125 SSSSAVRVRA-DFDPNIPIDESSWSSLELCEKLQAWYPMSKTLAEKAAWDYCKENGIDLV 183
SS SAV +P + +DE+ +S+ +LC + + WY +SKTLAE AAW + EN ID+V
Sbjct: 358 SSISAVVFDTRPKNPGVIVDETWFSNPDLCRESKLWYTLSKTLAEAAAWKFVNENSIDMV 537
Query: 184 TILPSFIIGPNLPTDLCSTASDVLGLFKGETEKFQWHGRMGYVHIDDVALCHILLYENKA 243
I P+ + GP L +L + +L L G + G+ ++ DVA HIL YE +
Sbjct: 538 AINPTMVAGPLLQPELNGSVEPILNLISGIPFP---NKAYGWCNVKDVANAHILAYETAS 708
Query: 244 SDGRYLCSSKIMDNDDLVGMLANRYPGFPIPKRFKKLDRPH---YELNTGKLESLGFKFK 300
+ GRY + +++ +L +L + YP I + + D P+ Y+++ K +SLG +F
Sbjct: 709 ASGRYCLAERVVHYSELAMILRDLYPTLQISDKCED-DGPYMPTYQISKEKAKSLGIEFT 885
Query: 301 SVEEMFDDCFASFVEQ 316
S+E + SF E+
Sbjct: 886 SLEVTLKETVESFREK 933
>TC87904 similar to GP|5852933|gb|AAD54273.1| dihydroflavonol-4-reductase
DFR1 {Glycine max}, partial (91%)
Length = 1244
Score = 222 bits (565), Expect = 2e-58
Identities = 121/318 (38%), Positives = 190/318 (59%), Gaps = 6/318 (1%)
Frame = +3
Query: 8 TGASGFLASWLIKRLLLSGYHVIGTVRDLGKKQKVEHLWKLEGATERLKLVQADLMEENS 67
TGASGF+ SWL+ RL+ GY V TVRD +KV HL +L GA +L L +ADL EE S
Sbjct: 42 TGASGFIGSWLVMRLMERGYMVRATVRDPENLKKVSHLLELPGAKGKLSLWKADLGEEGS 221
Query: 68 FDNAIMGCKGVFHIASPVLNHISNDPKAEILEPAVQGTLNVLRSCRKNPALVRVVLASSS 127
FD AI GC GVFH+A+P ++ S DP+ E+++P ++G L+++++C K + R + SS+
Sbjct: 222 FDEAIKGCTGVFHVATP-MDFESKDPENEMIKPTIKGVLDIMKACLKAKTVRRFIFTSSA 398
Query: 128 SAVRVRADFDPNIPIDESSWSSLELCE--KLQAW-YPMSKTLAEKAAWDYCKENGIDLVT 184
+ V D P DES WS +E C K+ W Y +SKTLAE+ AW + KE+ +D +T
Sbjct: 399 GTLNVTEDQKP--LWDESCWSDVEFCRRVKMTGWMYFVSKTLAEQEAWKFAKEHNMDFIT 572
Query: 185 ILPSFIIGPNLPTDLCSTASDVLGLFKGETEKFQWHGRMGYVHIDDVALCHILLYENKAS 244
I+P ++GP L + + L G + + +VH+DD+ HI L+E+
Sbjct: 573 IIPPLVVGPFLIPTMPPSLITALSPITGNEAHYSIIKQGQFVHLDDLCEAHIFLFEHMEV 752
Query: 245 DGRYLCSSKIMDNDDLVGMLANRYPGFPIPKRFKKL--DRPHYELNTGKLESLGFKFK-S 301
+GRYLCS+ + D+ ++ +YP + IP +F + + ++ K++ LGF+FK S
Sbjct: 753 EGRYLCSACEANIHDIAKLINTKYPEYNIPTKFNNIPDELELVRFSSKKIKDLGFEFKYS 932
Query: 302 VEEMFDDCFASFVEQGHL 319
+E+M+ + + +E+G L
Sbjct: 933 LEDMYTEAIDTCIEKGLL 986
>TC83473 similar to GP|5852933|gb|AAD54273.1| dihydroflavonol-4-reductase
DFR1 {Glycine max}, partial (96%)
Length = 1238
Score = 150 bits (378), Expect(2) = 7e-57
Identities = 80/170 (47%), Positives = 111/170 (65%), Gaps = 3/170 (1%)
Frame = +3
Query: 5 VCVTGASGFLASWLIKRLLLSGYHVIGTVRDLGKKQKVEHLWKLEGATERLKLVQADLME 64
VCVTGASGF+ SWL+ RL+ GY V TVRD +KV+HL +L GA +L L +ADL E
Sbjct: 69 VCVTGASGFIGSWLVMRLMERGYTVRATVRDPDNMKKVKHLLELPGANSKLSLWKADLGE 248
Query: 65 ENSFDNAIMGCKGVFHIASPVLNHISNDPKAEILEPAVQGTLNVLRSCRKNPALVRVVLA 124
E SFD AI GC GVFH+A+P ++ S DP+ E++ P + G L+++++C+K + R+V
Sbjct: 249 EGSFDEAIKGCTGVFHVATP-MDFESKDPEKEVINPTINGLLDIMKACKKAKTVRRLVFT 425
Query: 125 SSSSAVRVRADFDPNIPIDESSWSSLELCE--KLQAW-YPMSKTLAEKAA 171
SS+ + V N IDE+ WS +E C K+ W Y +SKTLAE+ A
Sbjct: 426 SSAGTLDVTE--QQNSVIDETCWSDVEFCRRVKMTGWMYFVSKTLAEQEA 569
Score = 88.6 bits (218), Expect(2) = 7e-57
Identities = 48/156 (30%), Positives = 82/156 (51%), Gaps = 3/156 (1%)
Frame = +1
Query: 174 YCKENGIDLVTILPSFIIGPNLPTDLCSTASDVLGLFKGETEKFQWHGRMGYVHIDDVAL 233
+ KE+ ID V+I+P ++GP + + + L L G + + Y+H+DD+ L
Sbjct: 577 FSKEHNIDFVSIIPPLVVGPFIMPSMPPSLITALSLITGYEAHYSIIKQGQYIHLDDLCL 756
Query: 234 CHILLYENKASDGRYLCSSKIMDNDDLVGMLANRYPGFPIPKRFKKL--DRPHYELNTGK 291
HI L+EN + GRY+C S ++ ++ +YP F +P +FK + D + ++ K
Sbjct: 757 AHIFLFENPKAHGRYICCSHEATIHEVAKLINKKYPEFNVPTKFKDIPDDLEIIKFSSKK 936
Query: 292 LESLGFKFK-SVEEMFDDCFASFVEQGHLTTLPHQP 326
+ LGF FK S+E+MF + E+G L + P
Sbjct: 937 ITDLGFIFKYSLEDMFTGAIETCREKGLLPKVTETP 1044
>TC77268 weakly similar to GP|10177026|dbj|BAB10264. dihydroflavonol
4-reductase-like {Arabidopsis thaliana}, partial (93%)
Length = 1240
Score = 214 bits (546), Expect = 3e-56
Identities = 122/327 (37%), Positives = 179/327 (54%), Gaps = 10/327 (3%)
Frame = +2
Query: 1 MEHKVCVTGASGFLASWLIKRLLLSGYHVIGTVRDLGKKQKVEHLWKLEGATERLKLVQA 60
M VCVTGASG + SW+++ LL GY V T++DL + + +HL +EGA RLK +
Sbjct: 32 MSKTVCVTGASGAIGSWVVRLLLERGYTVHATIQDLEDENETKHLEAMEGAKTRLKFFEM 211
Query: 61 DLMEENSFDNAIMGCKGVFHIASPVLNHISNDPKAEILEPAVQGTLNVLRSCRKNPALVR 120
DL+ +S A+ GC GV H+A P + DP+ +ILEPA+QGT+NVL+ K + R
Sbjct: 212 DLLNSDSIAAAVKGCAGVIHLACPNIIGEVKDPEKQILEPAIQGTVNVLK-VAKEAGVER 388
Query: 121 VVLASSSSAVRVRADFDPNIPIDESSWSSLELCEKLQAWYPMSKTLAEKAAWDYCKENGI 180
VV SS SA+ + + E W+ LE C++ + +YP++KTLAEKA W++ KE G
Sbjct: 389 VVATSSISAIIPSPSWPADKIKAEDCWTDLEYCKEKKLYYPIAKTLAEKAGWEFAKETGF 568
Query: 181 DLVTILPSFIIGPNLPTDLCSTASDVLGLFKGETEKFQWHGRMGYVHIDDVALCHILLYE 240
D+V I P +GP +P + S+ + + G+ KG+ E ++ MG H D+AL HIL +E
Sbjct: 569 DVVMINPGTALGPLIPPRINSSMAVLAGVLKGDKETYE-DFFMGMAHFKDIALAHILGFE 745
Query: 241 NKASDGRYLCSSKIMDNDDLVGMLANRYPGFPIPK----------RFKKLDRPHYELNTG 290
K + GR+LC I D V ++A YP + + K R K +
Sbjct: 746 QKKASGRHLCVEAIRHYSDFVNLVAELYPEYNVAKIPTDTQPGLLRAKNASK-------- 901
Query: 291 KLESLGFKFKSVEEMFDDCFASFVEQG 317
KL LG +F EE+ D +G
Sbjct: 902 KLIELGLEFTPAEEIIKDAVECLKSRG 982
>TC90858 anthocyanidin reductase [Medicago truncatula]
Length = 1181
Score = 209 bits (532), Expect = 1e-54
Identities = 124/326 (38%), Positives = 185/326 (56%), Gaps = 12/326 (3%)
Frame = +2
Query: 4 KVCVTGASGFLASWLIKRLLLSGYHVIGTVRDLGKKQKVEHLWKLEGATERLKLVQADLM 63
K CV G +GF+AS LIK+LL GY V TVRDL K HL L+ E L L +A+L
Sbjct: 77 KACVIGGTGFVASLLIKQLLEKGYAVNTTVRDLDSANKTSHLIALQSLGE-LNLFKAELT 253
Query: 64 EENSFDNAIMGCKGVFHIASPVLNHISNDPKAEILEPAVQGTLNVLRSCRKNPALVRVVL 123
E FD I GC+ VF +A+PV N S DP+ ++++PA++G LNVL++C + + RV+L
Sbjct: 254 IEEDFDAPISGCELVFQLATPV-NFASQDPENDMIKPAIKGVLNVLKACVRAKEVKRVIL 430
Query: 124 ASSSSAVRVRADFDPNIPIDESSWSSLELCE--KLQAW-YPMSKTLAEKAAWDYCKENGI 180
SS++AV + +DE++WS +E K W YP+SK LAEKAAW + +EN I
Sbjct: 431 TSSAAAVTINELEGTGHVMDETNWSDVEFLNTAKPPTWGYPVSKVLAEKAAWKFAEENNI 610
Query: 181 DLVTILPSFIIGPNLPTDLCSTASDVLGLFKGE------TEKFQW-HGRMGYVHIDDVAL 233
DL+T++P+ IGP+L D+ S+ + + L G + Q+ G + H++D+
Sbjct: 611 DLITVIPTLTIGPSLTQDIPSSVAMGMSLLTGNDFLINALKGMQFLSGSISITHVEDICR 790
Query: 234 CHILLYENKASDGRYLCSSKIMDNDDLVGMLANRYPGFPIPKRFKKL-DRPHYELNTGKL 292
HI + E +++ GRY+C + +L L+ RYP + +P F + +++GKL
Sbjct: 791 AHIFVAEKESTSGRYICCAHNTSVPELAKFLSKRYPQYKVPTEFDDFPSKAKLIISSGKL 970
Query: 293 ESLGFKFK-SVEEMFDDCFASFVEQG 317
GF FK S+ E FD QG
Sbjct: 971 IKEGFSFKHSIAETFDQTVEYLKTQG 1048
>TC77308 PIR|S66262|S66262 vestitone reductase - alfalfa, complete
Length = 1207
Score = 198 bits (503), Expect = 3e-51
Identities = 127/326 (38%), Positives = 187/326 (56%), Gaps = 10/326 (3%)
Frame = +3
Query: 4 KVCVTGASGFLASWLIKRLLLSGYHVIGTVR-DLGKKQKVEHLWKLEGATERLKLVQADL 62
+VCVTG +GFL SW+IK LL +GY V T+R D +K+ V L L GA+E+L ADL
Sbjct: 90 RVCVTGGTGFLGSWIIKSLLENGYSVNTTIRADPERKRDVRFLTNLPGASEKLHFFNADL 269
Query: 63 MEENSFDNAIMGCKGVFHIASPVLNHISNDPKAEILEPAVQGTLNVLRSCRKNPALVRVV 122
+SF AI GC G+FH ASP+ + ++P+ + + V G L +L++C + + R +
Sbjct: 270 SNPDSFAAAIEGCVGIFHTASPI-DFAVSEPEEIVTKRTVDGALGILKACVNSKTVKRFI 446
Query: 123 LASSSSAVRVRADFDPNIPIDESSWSSLELCEKLQAW---YPMSKTLAEKAAWDYCKENG 179
SS SAV D ++ +DES WS ++L ++ + Y +SKTLAEKA ++ ++NG
Sbjct: 447 YTSSGSAVSFNGK-DKDV-LDESDWSDVDLLRSVKPFGWNYAVSKTLAEKAVLEFGEQNG 620
Query: 180 IDLVTILPSFIIGPNLPTDLCSTASDVLGLFKGETEKFQWHGRMGYVHIDDVALCHILLY 239
ID+VT++ FI G + L + L L G+ E+ R VH+DDVA HI L
Sbjct: 621 IDVVTLILPFIGGRFVCPKLPDSIEKALVLVLGKKEQI-GVTRFHMVHVDDVARAHIYLL 797
Query: 240 ENKASDGRYLCSSKIMDNDDLVGMLANRYPGFPI-----PKRFKKLDRPHYELNTGKLES 294
EN GRY CS I+ +++ +L+ +YP + I K K P +LNT KL
Sbjct: 798 ENSVPGGRYNCSPFIVPIEEMSQLLSAKYPEYQILTVDELKEIKGARLP--DLNTKKLVD 971
Query: 295 LGFKFK-SVEEMFDDCFASFVEQGHL 319
GF+FK ++E+MFDD E+G+L
Sbjct: 972 AGFEFKYTIEDMFDDAIQCCKEKGYL 1049
>TC78732 weakly similar to PIR|C84436|C84436 probable cinnamoyl-CoA
reductase [imported] - Arabidopsis thaliana, partial
(86%)
Length = 1198
Score = 193 bits (491), Expect = 7e-50
Identities = 116/325 (35%), Positives = 175/325 (53%), Gaps = 7/325 (2%)
Frame = +1
Query: 2 EHKVCVTGASGFLASWLIKRLLLSGYHVIGTVRDLGKKQKVEHLWKLEG-ATERLKLVQA 60
E VCVTGA+GF+ SWL+ LL + HL+ L A R+ +
Sbjct: 61 EEVVCVTGANGFIGSWLVHTLLSKQNPHYKIHATIFPNSDPSHLFTLHPEAQSRITIFPV 240
Query: 61 DLMEENSFDNAIMGCKGVFHIASPVLNHISNDPKAEILEPAVQGTLNVLRSCRKNPALVR 120
++++ + NAI GC GVFH+ASP DP+ E+LEPAVQGTLNVL + K + R
Sbjct: 241 NILDSTAVSNAINGCSGVFHVASPCTLEDPTDPQKELLEPAVQGTLNVLEA-SKRAGVKR 417
Query: 121 VVLASSSSAVRVRADFDPNIPIDESSWSSLELCEKLQAWYPMSKTLAEKAAWDYC-KENG 179
VVL SS SA+ ++ N IDE SW+ +E C+ WYP+SKT AEK AW++C K +G
Sbjct: 418 VVLTSSISAMVPNPNWPENKAIDEGSWTDVEYCKLRGKWYPVSKTEAEKVAWEFCEKHSG 597
Query: 180 IDLVTILPSFIIGPNLPTDLCSTASDVLGLFKGE--TEKFQWHGRMGYVHIDDVALCHIL 237
+D+V + P +GP L + ++++ + L GE T++ W +G VH+ DVA H+L
Sbjct: 598 VDVVAVHPGTCLGPLLQNQMNASSAVLQRLMMGEKDTQECYW---LGAVHVKDVARAHVL 768
Query: 238 LYENKASDGRYLCSSKIMDNDDLVGMLANRYPGFPIPKRFKKLDRP---HYELNTGKLES 294
+YE + GRYLC + I +++ Y +PI F +P ++ +L
Sbjct: 769 VYETPTAAGRYLCVNGIYQFSSFAKIVSELYHDYPI-HSFPNETQPGLTPFKEAAKRLID 945
Query: 295 LGFKFKSVEEMFDDCFASFVEQGHL 319
LG F +++ + S + +G L
Sbjct: 946 LGLVFTPIQDAIREAAESLMAKGFL 1020
>BE997470 similar to PIR|D84747|D84 probable cinnamoyl-CoA reductase
[imported] - Arabidopsis thaliana, partial (44%)
Length = 807
Score = 182 bits (461), Expect = 2e-46
Identities = 104/246 (42%), Positives = 151/246 (61%), Gaps = 8/246 (3%)
Frame = +3
Query: 4 KVCVTGASGFLASWLIKRLLLSGYHVIGTVRDLGKKQKVEHLWKLEGATERLKLVQADLM 63
KVCVTGA GF+ASWL+K LL GY V GTVR+ G K EHL KLE A+E L L +AD++
Sbjct: 90 KVCVTGAGGFVASWLVKLLLSKGYFVHGTVREPGSP-KYEHLLKLEKASENLTLFKADIL 266
Query: 64 EENSFDNAIMGCKGVFHIASPVLNHISNDPK----AEILEPAVQGTLNVLRSCRKNPALV 119
+ S +AI+GC VFH+ASPV + + +P+ E++EPAV+GT NVL +C K +
Sbjct: 267 DYESVYSAIVGCSAVFHVASPVPSTVVPNPELNLQVEVIEPAVKGTANVLEACLK-ANVE 443
Query: 120 RVVLASSSSAVRVRADFDPNIPIDESSWSSLELCEKLQAWYPMSKTLAEKAAWDYCKENG 179
RVV SS++AV + + + IDES WS + C+ Q WY +KT AE+ A+++ K G
Sbjct: 444 RVVFVSSAAAVAINPNLPKDKAIDESCWSDKDYCKNTQNWYCYAKTEAEEQAFNFAKRTG 623
Query: 180 IDLVTILPSFIIGPNLPTDLCSTASDVLGLFK----GETEKFQWHGRMGYVHIDDVALCH 235
+++VTI P+ ++GP L + S++ ++ + K K +W V + DV
Sbjct: 624 LNVVTICPTLVLGPILQSTTNSSSLALIKILKEGHDSLENKLRW-----IVDVRDVVNAI 788
Query: 236 ILLYEN 241
+L YEN
Sbjct: 789 LLAYEN 806
>AW696839 similar to PIR|T16995|T16 probable cinnamyl-alcohol dehydrogenase
(EC 1.1.1.195) - apple tree, partial (56%)
Length = 579
Score = 133 bits (335), Expect(3) = 8e-43
Identities = 65/115 (56%), Positives = 88/115 (76%)
Frame = +3
Query: 16 SWLIKRLLLSGYHVIGTVRDLGKKQKVEHLWKLEGATERLKLVQADLMEENSFDNAIMGC 75
SWL++ LL GY V TVRD +K+ HL KLEGA ERL+L +A+L+E+ +FD+A+ GC
Sbjct: 66 SWLVRYLLHRGYTVKATVRDPSDPKKINHLVKLEGAKERLQLFKANLLEQGAFDSAVQGC 245
Query: 76 KGVFHIASPVLNHISNDPKAEILEPAVQGTLNVLRSCRKNPALVRVVLASSSSAV 130
GVFH ASP +H+ DP+AE+++PA+ GTLNVL+SC K+P L RVVL SS++AV
Sbjct: 246 HGVFHTASPFYHHV-KDPQAELIDPALNGTLNVLKSCAKSPLLKRVVLTSSAAAV 407
Score = 54.3 bits (129), Expect(3) = 8e-43
Identities = 23/42 (54%), Positives = 31/42 (73%)
Frame = +1
Query: 144 ESSWSSLELCEKLQAWYPMSKTLAEKAAWDYCKENGIDLVTI 185
E+ ++ + C KL WY +SKTLAE+AAW + KEN ID+VTI
Sbjct: 451 ETWFTDADFCAKLNLWYAVSKTLAEEAAWKFVKENNIDMVTI 576
Score = 24.3 bits (51), Expect(3) = 8e-43
Identities = 9/11 (81%), Positives = 11/11 (99%)
Frame = +2
Query: 5 VCVTGASGFLA 15
VCVTGASG++A
Sbjct: 32 VCVTGASGYIA 64
>AW559294 similar to GP|21594240|gb cinnamyl-alcohol dehydrogenase-like
protein {Arabidopsis thaliana}, partial (43%)
Length = 531
Score = 149 bits (377), Expect = 1e-36
Identities = 73/122 (59%), Positives = 95/122 (77%)
Frame = +2
Query: 5 VCVTGASGFLASWLIKRLLLSGYHVIGTVRDLGKKQKVEHLWKLEGATERLKLVQADLME 64
VCVTGASGF+ASW++K LL GY V TVRD KV+HL KL+GA ERL+L +ADL+E
Sbjct: 92 VCVTGASGFIASWVVKFLLQRGYTVRATVRDPSNSNKVDHLLKLDGAKERLQLFKADLLE 271
Query: 65 ENSFDNAIMGCKGVFHIASPVLNHISNDPKAEILEPAVQGTLNVLRSCRKNPALVRVVLA 124
E SFD+ I GC GVFH ASPV + + DP+ ++++PAV+GTLNV++SC K+P++ RVVL
Sbjct: 272 EGSFDSVIQGCHGVFHTASPV-HFVVTDPQTQLIDPAVKGTLNVVKSCAKSPSVQRVVLT 448
Query: 125 SS 126
SS
Sbjct: 449 SS 454
>TC79596 similar to GP|21594240|gb|AAM65984.1 cinnamyl-alcohol
dehydrogenase-like protein {Arabidopsis thaliana},
partial (34%)
Length = 531
Score = 108 bits (270), Expect(2) = 3e-30
Identities = 52/81 (64%), Positives = 63/81 (77%)
Frame = +2
Query: 5 VCVTGASGFLASWLIKRLLLSGYHVIGTVRDLGKKQKVEHLWKLEGATERLKLVQADLME 64
VCVTGASGF+ASW++K LL GY V TVRD +KV+HL KL+GA ERL+L +ADL+E
Sbjct: 140 VCVTGASGFIASWIVKFLLQRGYTVRATVRDPSNPKKVDHLLKLDGAKERLQLFKADLLE 319
Query: 65 ENSFDNAIMGCKGVFHIASPV 85
E SFD+ + GC GVFH ASPV
Sbjct: 320 EGSFDSVVEGCDGVFHTASPV 382
Score = 40.8 bits (94), Expect(2) = 3e-30
Identities = 15/28 (53%), Positives = 25/28 (88%)
Frame = +3
Query: 91 NDPKAEILEPAVQGTLNVLRSCRKNPAL 118
NDP+ E+++PA++GTLNVL+SC K+ ++
Sbjct: 396 NDPQVELIDPALKGTLNVLKSCAKSTSV 479
>TC78040 similar to GP|23495824|dbj|BAC20034. putative NADPH HC toxin
reductase {Oryza sativa (japonica cultivar-group)},
partial (33%)
Length = 1151
Score = 127 bits (318), Expect = 8e-30
Identities = 105/347 (30%), Positives = 174/347 (49%), Gaps = 21/347 (6%)
Frame = +1
Query: 4 KVCVTGASGFLASWLIKRLLLSGYHVIGTVRDLGKKQKVEHLWKLEGATERLKLVQADLM 63
KVCVTGASG++AS LI +LL GY V T+RDL + KV L + ++L L QAD+
Sbjct: 76 KVCVTGASGYIASLLINKLLAKGYTVHATLRDLKDESKVGLLKSFPQSQDKLVLFQADIY 255
Query: 64 EENSFDNAIMGCKGVFHIASPVLNHISNDPKAEILEPAVQGTLNVLRSCRKNPALVRVVL 123
F+ AI GC+ VFH+A+P+++ ++ K +I E ++ G+ ++ C+K + R++
Sbjct: 256 NSVDFEPAIKGCEFVFHVATPLIHEPASQFK-DITEASLAGSESIAMYCKKAGTVKRLIY 432
Query: 124 ASS--SSAVRVRADFDPNIPIDESSWSSLE------LCEKLQAWYPMSKTLAEKAAWDYC 175
S S++ R N +DE+ W+ L + Y SKT+ EK C
Sbjct: 433 TGSVVSASPRRVDGIGFNDVMDETCWTPLNDSLAYLYHDAYLKDYIYSKTVTEKYMLS-C 609
Query: 176 --KENG--IDLVTILPSFIIGPNL----PTDLCSTASDVLGLFKG-ETEKF--QWHGRMG 224
ENG +++VT+L + G L P + S + G ++ +F ++ G++
Sbjct: 610 GNNENGGRLEVVTLLCGAVGGDTLQSFTPGSVAICISHITENAMGRKSLQFVQEFLGKIP 789
Query: 225 YVHIDDVALCHILLYENKAS-DGRYLCSSKIMDNDDLVGMLANRYPGFPIPKRFKKLDRP 283
VH+DDV HI E+ +S +GR+LC+S + ++ YP F + + + +
Sbjct: 790 LVHVDDVCEAHIFCMESTSSINGRFLCASSYVSLKEIANHYVLHYPKFTVNQEYADGPKK 969
Query: 284 HYELNTGKLESLGFKFK-SVEEMFDDCFASFVEQGHLTTLPHQPRVI 329
+ + KL GF +K + + DDC G L + P+ P I
Sbjct: 970 DMKWGSTKLCDKGFVYKYDAKMILDDCVKCARRMGDL*S-PYPPCTI 1107
>TC81862 weakly similar to PIR|T05749|T05749 hypothetical protein M4I22.60 -
Arabidopsis thaliana, partial (43%)
Length = 755
Score = 76.6 bits (187), Expect(2) = 2e-29
Identities = 38/80 (47%), Positives = 52/80 (64%)
Frame = +1
Query: 4 KVCVTGASGFLASWLIKRLLLSGYHVIGTVRDLGKKQKVEHLWKLEGATERLKLVQADLM 63
K CVTGA+G++ SWL++ LL G V TVRD K + LWK ++L+ +ADL
Sbjct: 91 KYCVTGATGYIGSWLVEALLQRGCTVHATVRDPEKSLHLLSLWK---GGDQLRFFRADLH 261
Query: 64 EENSFDNAIMGCKGVFHIAS 83
EE SF+ A+ GC GVFH+A+
Sbjct: 262 EEGSFEEAVKGCDGVFHVAA 321
Score = 70.1 bits (170), Expect(2) = 2e-29
Identities = 42/110 (38%), Positives = 65/110 (58%), Gaps = 4/110 (3%)
Frame = +2
Query: 88 HISNDPKAEILEPAVQGTLNVLRSCRKNPALVRVVLASSSSAVRVRADFDPNIPI-DESS 146
+I N + I++PA++GT+N+L+SC K+ ++ RVV SS S + + PI DES
Sbjct: 353 NIENFVQTNIIDPAIEGTVNLLKSCLKSNSVKRVVFTSSISTITAKDSNGKWKPIVDESC 532
Query: 147 WSSLELCEKLQ--AW-YPMSKTLAEKAAWDYCKENGIDLVTILPSFIIGP 193
+ Q W Y +SK L E+AA+ + KENGIDLV+++ S + P
Sbjct: 533 QIQTDTVWNTQPSGWVYALSKLLTEEAAFKFAKENGIDLVSVITSTVCRP 682
>TC77866 similar to GP|18266655|gb|AAL67601.1 putative cinnamoyl-CoA
reductase {Oryza sativa}, partial (58%)
Length = 1300
Score = 118 bits (296), Expect = 3e-27
Identities = 82/266 (30%), Positives = 137/266 (50%), Gaps = 1/266 (0%)
Frame = +1
Query: 3 HKVCVTGASGFLASWLIKRLLLSGYHVIGTVRDLGKKQKVEHLWKLEGATERLKLVQADL 62
H VCV ASG L L++RLL GY V +++ G+ + +++LK+ ++D
Sbjct: 235 HTVCVMDASGQLGFSLVQRLLQRGYTVHASIQQYGENP----FNGISADSDKLKIFRSDP 402
Query: 63 MEENSFDNAIMGCKGVFHIASPVLNHISNDPKAEILEPAVQGTLNVLRSCRKNPALVRVV 122
+ +S +A+ GC G+F+ P L+ + D +E V+ NVL +C + + +VV
Sbjct: 403 FDYHSIIDALRGCSGLFYSFEPPLDQPNYDEYMADVE--VRAAHNVLEACAQTETIDKVV 576
Query: 123 LASSSSAVRVRADFDPNIP-IDESSWSSLELCEKLQAWYPMSKTLAEKAAWDYCKENGID 181
SS++AV R D P +DE WS + C K + W+ MSKTLAEK A+ + G++
Sbjct: 577 FTSSATAVVWREDRKTIEPDLDEIHWSDVNFCRKFKLWHGMSKTLAEKTAFALAMDRGVN 756
Query: 182 LVTILPSFIIGPNLPTDLCSTASDVLGLFKGETEKFQWHGRMGYVHIDDVALCHILLYEN 241
+V+I ++ +L S +G E ++ G V + + HI +YE+
Sbjct: 757 MVSINAGLLMTHDL--------SIKHPYLRGAAEMYE-DGVFVTVDLGFLVDTHICVYED 909
Query: 242 KASDGRYLCSSKIMDNDDLVGMLANR 267
+S GRYLC + I++ D LA++
Sbjct: 910 VSSYGRYLCFNHIINTQDDAVQLAHK 987
>AL382470 similar to PIR|S66262|S66 vestitone reductase - alfalfa, partial
(48%)
Length = 550
Score = 114 bits (284), Expect = 7e-26
Identities = 63/160 (39%), Positives = 100/160 (62%), Gaps = 4/160 (2%)
Frame = -3
Query: 4 KVCVTGASGFLASWLIKRLLLSGYHVIGTVRD--LGKKQKVEHLWKLEGATERLKLVQAD 61
+VCVTG +GF+ SW+IKRLL GY V TVR G+K+ V +L L A++ L++ AD
Sbjct: 473 RVCVTGGTGFIGSWIIKRLLEDGYTVNTTVRSNPAGQKKDVSYLTNLPNASQNLQIFNAD 294
Query: 62 LMEENSFDNAIMGCKGVFHIASPVLNHISNDPKAEILEPAVQGTLNVLRSCRKNPALVRV 121
L SFD AI GC G+FH A+P+ + N+ + + + + G L +L++C+ + + RV
Sbjct: 293 LCNPESFDAAIEGCIGIFHTATPI-DFEENEREEIVTKRTIDGALGILKACKNSKTVKRV 117
Query: 122 VLASSSSAVRVRADFDPNIPIDESSWSSLELCEKLQ--AW 159
+ SS+SAV ++ D + ++ +DES WS + + L+ AW
Sbjct: 116 IYTSSASAVYMQ-DKEEDV-MDESYWSDVNILRNLKPFAW 3
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.321 0.138 0.422
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,507,947
Number of Sequences: 36976
Number of extensions: 166990
Number of successful extensions: 835
Number of sequences better than 10.0: 82
Number of HSP's better than 10.0 without gapping: 772
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 778
length of query: 330
length of database: 9,014,727
effective HSP length: 96
effective length of query: 234
effective length of database: 5,465,031
effective search space: 1278817254
effective search space used: 1278817254
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 58 (26.9 bits)
Medicago: description of AC138130.14


