

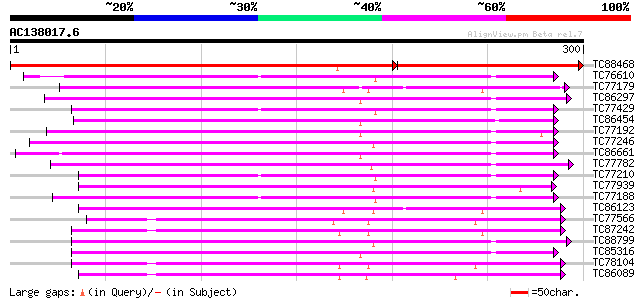
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC138017.6 - phase: 0
(300 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC88468 homologue to PIR|T10690|T10690 serine/threonine-specific... 403 e-166
TC76610 similar to PIR|T50802|T50802 serine/threonine protein ki... 179 9e-46
TC77179 similar to GP|2665890|gb|AAB88537.1| calcium-dependent p... 174 4e-44
TC86297 similar to GP|15912283|gb|AAL08275.1 At1g30270/F12P21_6 ... 169 2e-42
TC77429 similar to GP|15215664|gb|AAK91377.1 AT5g25110/T11H3_120... 163 7e-41
TC86454 homologue to GP|4567091|gb|AAD23582.1| SNF-1-like serine... 161 2e-40
TC77192 similar to SP|P42818|KPK1_ARATH Serine/threonine-protein... 160 7e-40
TC77246 similar to PIR|C71408|C71408 probable protein kinase - A... 159 1e-39
TC86661 similar to GP|9798599|dbj|BAB11737.1 serine/threonine pr... 159 2e-39
TC77782 similar to GP|13448037|gb|AAK26845.1 SOS2-like protein k... 157 5e-39
TC77210 similar to GP|21954717|gb|AAM83095.1 SOS2-like protein k... 154 5e-38
TC77939 similar to GP|14194167|gb|AAK56278.1 At2g26980/T20P8.3 {... 153 9e-38
TC77188 similar to PIR|T04862|T04862 probable serine/threonine-s... 150 6e-37
TC86123 homologue to PIR|S71770|S71770 calcium-dependent protein... 147 4e-36
TC77566 similar to GP|310580|gb|AAA34017.1|| protein kinase 2 {G... 147 4e-36
TC87242 homologue to PIR|S56716|S56716 protein kinase SPK-3 (EC ... 147 4e-36
TC88799 similar to PIR|C71408|C71408 probable protein kinase - A... 145 2e-35
TC85316 weakly similar to PIR|E84707|E84707 probable protein kin... 145 2e-35
TC78104 homologue to GP|19568098|gb|AAL89456.1 osmotic stress-ac... 144 4e-35
TC86089 similar to PIR|S71172|S71172 protein kinase 41K (EC 2.7... 144 5e-35
>TC88468 homologue to PIR|T10690|T10690 serine/threonine-specific protein
kinase homolog T16I18.40 - Arabidopsis thaliana, partial
(92%)
Length = 1591
Score = 403 bits (1035), Expect(2) = e-166
Identities = 203/235 (86%), Positives = 203/235 (86%), Gaps = 32/235 (13%)
Frame = +1
Query: 1 MAIATETQPQPHQQHHTASSEVSGSAKDQRRWILNDFDIGKPLGRGKFGHVYLAREKTSN 60
MAIATETQPQPHQQHHTASSEVSGSAKDQRRWILNDFDIGKPLGRGKFGHVYLAREKTSN
Sbjct: 91 MAIATETQPQPHQQHHTASSEVSGSAKDQRRWILNDFDIGKPLGRGKFGHVYLAREKTSN 270
Query: 61 HIVALKVLFKSQLQQSQVEHQLRREVEIQSHLRHPHILRLYGYFYDQKRVYLILEYAPKG 120
HIVALKVLFKSQLQQSQVEHQLRREVEIQSHLRHPHILRLYGYFYDQKRVYLILEYAPKG
Sbjct: 271 HIVALKVLFKSQLQQSQVEHQLRREVEIQSHLRHPHILRLYGYFYDQKRVYLILEYAPKG 450
Query: 121 ELYKELQKCKYFSERRAATYVASLARALIYCHGKHVIHRDIKPENLLIGA---------- 170
ELYKELQKCKYFSERRAATYVASLARALIYCHGKHVIHRDIKPENLLIGA
Sbjct: 451 ELYKELQKCKYFSERRAATYVASLARALIYCHGKHVIHRDIKPENLLIGAQVIPHT*TIV 630
Query: 171 ----------------------QGELKIADFGWSVHTFNRRRTMCGTLDYLPPEM 203
QGELKIADFGWSVHTFNRRRTMCGTLDYLPPEM
Sbjct: 631 SFMVLLLVVFNLC*SCENLLHYQGELKIADFGWSVHTFNRRRTMCGTLDYLPPEM 795
Score = 200 bits (508), Expect(2) = e-166
Identities = 97/97 (100%), Positives = 97/97 (100%)
Frame = +3
Query: 204 VESVEHDASVDIWSLGVLCYEFLYGVPPFEAKEHSDTYRRIIQVDLKFPPKPIVSSAAKD 263
VESVEHDASVDIWSLGVLCYEFLYGVPPFEAKEHSDTYRRIIQVDLKFPPKPIVSSAAKD
Sbjct: 885 VESVEHDASVDIWSLGVLCYEFLYGVPPFEAKEHSDTYRRIIQVDLKFPPKPIVSSAAKD 1064
Query: 264 LISQMLVKDSSERLPLHKLLEHPWIVQNAEPSGIYRS 300
LISQMLVKDSSERLPLHKLLEHPWIVQNAEPSGIYRS
Sbjct: 1065LISQMLVKDSSERLPLHKLLEHPWIVQNAEPSGIYRS 1175
>TC76610 similar to PIR|T50802|T50802 serine/threonine protein kinase-like
protein - Arabidopsis thaliana, partial (73%)
Length = 1799
Score = 179 bits (455), Expect = 9e-46
Identities = 101/286 (35%), Positives = 157/286 (54%), Gaps = 6/286 (2%)
Frame = +2
Query: 8 QPQPHQQHHTASSEVSGSAKDQRRWILNDFDIGKPLGRGKFGHVYLAREKTSNHIVALKV 67
QP P +QH +R + N ++IGK LG+G F VY R +N VA+KV
Sbjct: 275 QPLPMEQHR------------KRNILFNKYEIGKTLGQGNFAKVYHGRNIATNENVAIKV 418
Query: 68 LFKSQLQQSQVEHQLRREVEIQSHLRHPHILRLYGYFYDQKRVYLILEYAPKGELYKELQ 127
+ K +L++ ++ Q++REV + +RHPHI+ L ++ +V++++EY GEL+ ++
Sbjct: 419 IKKEKLKKERLMKQIKREVSVMRLVRHPHIVELKEVMANKAKVFMVVEYVKGGELFAKVA 598
Query: 128 KCKYFSERRAATYVASLARALIYCHGKHVIHRDIKPENLLIGAQGELKIADFGWSVHTFN 187
K K E A Y L A+ +CH + V HRD+KPENLL+ +LK++DFG S
Sbjct: 599 KGK-MEENIARKYFQQLISAVDFCHSRGVTHRDLKPENLLLDENEDLKVSDFGLSALPDQ 775
Query: 188 RRR-----TMCGTLDYLPPEMVESVEHDAS-VDIWSLGVLCYEFLYGVPPFEAKEHSDTY 241
RR T CGT Y+ PE+++ + +D S DIWS GV+ + L G PF+ + Y
Sbjct: 776 RRSDGMLLTPCGTPAYVAPEVLKKIGYDGSKADIWSCGVILFALLCGYLPFQGENVMRIY 955
Query: 242 RRIIQVDLKFPPKPIVSSAAKDLISQMLVKDSSERLPLHKLLEHPW 287
+ + D P +S AK LIS +LV D +R + +++ PW
Sbjct: 956 SKSFKADYVLP--EWISPGAKKLISNLLVVDPEKRFSIPDIMKDPW 1087
>TC77179 similar to GP|2665890|gb|AAB88537.1| calcium-dependent protein
kinase {Fragaria x ananassa}, partial (92%)
Length = 2243
Score = 174 bits (441), Expect = 4e-44
Identities = 104/276 (37%), Positives = 161/276 (57%), Gaps = 9/276 (3%)
Frame = +1
Query: 27 KDQRRWILNDFDIGKPLGRGKFGHVYLAREKTSNHIVALKVLFKSQLQQSQVEHQLRREV 86
K R IL +++G+ LGRG+FG YL +++ + +A K + K +L+ + +RREV
Sbjct: 364 KPTGREILQQYELGRELGRGEFGITYLCKDRETGEELACKSISKDKLRTAIDIEDVRREV 543
Query: 87 EIQSHL-RHPHILRLYGYFYDQKRVYLILEYAPKGELYKELQKCKYFSERRAATYVASLA 145
EI HL +HP+I+ L + D V+L++E GEL+ + +++ER AAT V ++
Sbjct: 544 EIMRHLPKHPNIVTLKDTYEDDDNVHLVMELCEGGELFDRIVAKGHYTERAAATVVKTIV 723
Query: 146 RALIYCHGKHVIHRDIKPENLLIGAQGE---LKIADFGWSVHTF---NRRRTMCGTLDYL 199
+ + CH V+HRD+KPEN L + E LK DFG S+ TF ++ + G+ Y+
Sbjct: 724 QVVQMCHEHGVMHRDLKPENFLFANKKETSPLKAIDFGLSI-TFKPGDKFNEIVGSPYYM 900
Query: 200 PPEMVESVEHDASVDIWSLGVLCYEFLYGVPPFEAKEHSDTYRRIIQ--VDLKFPPKPIV 257
PE+++ + +DIWS GV+ Y L G+PPF A+ + II+ +D K P P V
Sbjct: 901 APEVLKR-NYGPEIDIWSAGVILYILLCGIPPFWAETEQGIAQAIIRSVIDFKKEPWPKV 1077
Query: 258 SSAAKDLISQMLVKDSSERLPLHKLLEHPWIVQNAE 293
S AKDLI +ML D RL ++L+HPW+ QNA+
Sbjct: 1078SDNAKDLIKKMLDPDPKRRLTAQEVLDHPWL-QNAK 1182
>TC86297 similar to GP|15912283|gb|AAL08275.1 At1g30270/F12P21_6
{Arabidopsis thaliana}, partial (90%)
Length = 2043
Score = 169 bits (427), Expect = 2e-42
Identities = 87/282 (30%), Positives = 157/282 (54%), Gaps = 6/282 (2%)
Frame = +2
Query: 19 SSEVSGSAKDQRRWILNDFDIGKPLGRGKFGHVYLAREKTSNHIVALKVLFKSQLQQSQV 78
SS +SG + + R +++G+ LG G F V AR + VA+K+L K ++ + ++
Sbjct: 395 SSHISGGSSNGGRAKAGKYELGRTLGEGNFAKVKFARHIETGDHVAIKILDKEKILKHKM 574
Query: 79 EHQLRREVEIQSHLRHPHILRLYGYFYDQKRVYLILEYAPKGELYKELQKCKYFSERRAA 138
Q+++E+ +RHP+++R++ ++ ++++++E GEL+ ++ + E A
Sbjct: 575 IRQIKQEISTMKLIRHPNVIRMHEVIANRSKIFIVMELVTGGELFDKIARSGRLKEDEAR 754
Query: 139 TYVASLARALIYCHGKHVIHRDIKPENLLIGAQGELKIADFGWS-----VHTFNRRRTMC 193
Y L A+ YCH + V HRD+KPENLL+ G LK++DFG S V T C
Sbjct: 755 KYFQQLICAVDYCHSRGVCHRDLKPENLLLDTNGTLKVSDFGLSALPQQVREDGLLHTTC 934
Query: 194 GTLDYLPPEMVESVEHDASV-DIWSLGVLCYEFLYGVPPFEAKEHSDTYRRIIQVDLKFP 252
GT +Y+ PE++++ +D ++ D+WS GV+ + + G PFE Y++I + D P
Sbjct: 935 GTPNYVAPEVIQNKGYDGAIADLWSCGVILFVLMAGYLPFEEDNLVALYKKIFKADFTCP 1114
Query: 253 PKPIVSSAAKDLISQMLVKDSSERLPLHKLLEHPWIVQNAEP 294
P SS+AK LI ++L R+ + +++E+ W + +P
Sbjct: 1115--PWFSSSAKKLIKRILDPSPITRIKIAEVIENEWFKKGYKP 1234
>TC77429 similar to GP|15215664|gb|AAK91377.1 AT5g25110/T11H3_120
{Arabidopsis thaliana}, partial (79%)
Length = 1869
Score = 163 bits (413), Expect = 7e-41
Identities = 88/261 (33%), Positives = 147/261 (55%), Gaps = 6/261 (2%)
Frame = +3
Query: 33 ILNDFDIGKPLGRGKFGHVYLAREKTSNHIVALKVLFKSQLQQSQVEHQLRREVEIQSHL 92
+ +++G+ LG+G F VY A+E ++ VA+KV+ K ++++ + Q++RE+ + +
Sbjct: 258 LFGKYEMGRVLGKGTFAKVYYAKEISTGEGVAIKVVDKDKVKKEGMMEQIKREISVMRLV 437
Query: 93 RHPHILRLYGYFYDQKRVYLILEYAPKGELYKELQKCKYFSERRAATYVASLARALIYCH 152
+HP+I+ L + ++ ++EYA GEL++++ K K F E A Y L A+ YCH
Sbjct: 438 KHPNIVNLKEVMATKTKILFVMEYARGGELFEKVAKGK-FKEELARKYFQQLISAVDYCH 614
Query: 153 GKHVIHRDIKPENLLIGAQGELKIADFGWSVHTFNRRR-----TMCGTLDYLPPEMVESV 207
+ V HRD+KPENLL+ LK++DFG S + R+ T CGT Y+ PE+V
Sbjct: 615 SRGVSHRDLKPENLLLDENENLKVSDFGLSALPEHLRQDGLLHTQCGTPAYVAPEVVRKR 794
Query: 208 EHDA-SVDIWSLGVLCYEFLYGVPPFEAKEHSDTYRRIIQVDLKFPPKPIVSSAAKDLIS 266
+ D WS GV+ Y L G PF+ + Y ++ + + +FP P S +K LIS
Sbjct: 795 GYSGFKADTWSCGVILYALLAGFLPFQHENLISMYNKVFKEEYQFP--PWFSPESKRLIS 968
Query: 267 QMLVKDSSERLPLHKLLEHPW 287
++LV D R+ + ++ PW
Sbjct: 969 KILVADPERRITISSIMNVPW 1031
>TC86454 homologue to GP|4567091|gb|AAD23582.1| SNF-1-like serine/threonine
protein kinase {Glycine max}, partial (90%)
Length = 2194
Score = 161 bits (408), Expect = 2e-40
Identities = 85/257 (33%), Positives = 146/257 (56%), Gaps = 3/257 (1%)
Frame = +3
Query: 34 LNDFDIGKPLGRGKFGHVYLAREKTSNHIVALKVLFKSQLQQSQVEHQLRREVEIQSHLR 93
L ++ +GK LG G FG V +A + H VA+K+L + +++ ++E ++RRE++I
Sbjct: 105 LRNYKMGKTLGIGSFGKVKIAEHVLTGHKVAIKILNRRKIKNMEMEEKVRREIKILRLFM 284
Query: 94 HPHILRLYGYFYDQKRVYLILEYAPKGELYKELQKCKYFSERRAATYVASLARALIYCHG 153
H HI+RLY +Y+++EY GEL+ + + E A ++ + + YCH
Sbjct: 285 HHHIIRLYEVVETPTDIYVVMEYVKSGELFDYIVEKGRLQEDEARSFFQQIISGVEYCHR 464
Query: 154 KHVIHRDIKPENLLIGAQGELKIADFGWS--VHTFNRRRTMCGTLDYLPPEMVE-SVEHD 210
V+HRD+KPENLL+ ++ +KIADFG S + + +T CG+ +Y PE++ +
Sbjct: 465 NMVVHRDLKPENLLLDSKWSVKIADFGLSNIMRDGHFLKTSCGSPNYAAPEVISGKLYAG 644
Query: 211 ASVDIWSLGVLCYEFLYGVPPFEAKEHSDTYRRIIQVDLKFPPKPIVSSAAKDLISQMLV 270
VD+WS GV+ Y L G PF+ + + +++I P +S A+DLI ++LV
Sbjct: 645 PEVDVWSCGVILYALLCGTLPFDDENIPNLFKKIKGGIYTLPSH--LSPGARDLIPRLLV 818
Query: 271 KDSSERLPLHKLLEHPW 287
D +R+ + ++ +HPW
Sbjct: 819 VDPMKRMTIPEIRQHPW 869
>TC77192 similar to SP|P42818|KPK1_ARATH Serine/threonine-protein kinase
AtPK1/AtPK6 (EC 2.7.1.-). [Mouse-ear cress] {Arabidopsis
thaliana}, partial (76%)
Length = 1992
Score = 160 bits (404), Expect = 7e-40
Identities = 86/275 (31%), Positives = 152/275 (55%), Gaps = 7/275 (2%)
Frame = +3
Query: 20 SEVSGSAKDQRRWILNDFDIGKPLGRGKFGHVYLAREKTSNHIVALKVLFKSQLQQSQVE 79
++ G+ + RR + DF++ K +G+G F VY R+K ++ I A+KV+ K ++ +
Sbjct: 717 NDEDGNLMEIRRVGIEDFEVLKVVGQGAFAKVYQVRKKGTSEIYAMKVMRKDKIMEKNHA 896
Query: 80 HQLRREVEIQSHLRHPHILRLYGYFYDQKRVYLILEYAPKGELYKELQKCKYFSERRAAT 139
++ E EI + + HP +++L F + R+YL+L++ G L+ +L F E A
Sbjct: 897 EYMKAEREILTKIEHPFVVQLRYSFQTKYRLYLVLDFVNGGHLFFQLYHQGLFREDLARI 1076
Query: 140 YVASLARALIYCHGKHVIHRDIKPENLLIGAQGELKIADFGWS--VHTFNRRRTMCGTLD 197
Y A + A+ + H K ++HRD+KPEN+L+ A G + + DFG + R ++CGT +
Sbjct: 1077YAAEIVSAVSHLHSKGIMHRDLKPENILMDADGHVMLTDFGLAKKFEESTRSNSLCGTAE 1256
Query: 198 YLPPEMVESVEHDASVDIWSLGVLCYEFLYGVPPFEAKEHSDTYRRIIQVDLKFPPKPIV 257
Y+ PE++ HD + D WS+G+L +E L G PPF ++I++ +K P +
Sbjct: 1257YMAPEIILGKGHDKAADWWSVGILLFEMLTGKPPFCGGNCQKIQQKIVKDKIKLP--GFL 1430
Query: 258 SSAAKDLISQMLVKDSSERL-----PLHKLLEHPW 287
SS A L+ +L K++ +RL + ++ H W
Sbjct: 1431SSDAHALLKGLLNKEAPKRLGCGAKGIEEIKGHKW 1535
>TC77246 similar to PIR|C71408|C71408 probable protein kinase - Arabidopsis
thaliana, partial (65%)
Length = 1715
Score = 159 bits (402), Expect = 1e-39
Identities = 96/283 (33%), Positives = 149/283 (51%), Gaps = 6/283 (2%)
Frame = +3
Query: 11 PHQQHHTASSEVSGSAKDQRRWILNDFDIGKPLGRGKFGHVYLAREKTSNHIVALKVLFK 70
P++ S S R IL + + K LGRG F VY AR VA+KV+ K
Sbjct: 9 PNKMERRPQSPPSPPPSPPSRIILGKYQLTKFLGRGNFAKVYQARSLIDGSTVAVKVIDK 188
Query: 71 SQLQQSQVEHQLRREVEIQSHLR-HPHILRLYGYFYDQKRVYLILEYAPKGELYKELQKC 129
S+ + +E ++ RE++ L+ HP+IL+++ + ++YLI+++A GEL+ +L +
Sbjct: 189 SKTVDASMEPRIVREIDAMRRLQNHPNILKIHEVLATKTKIYLIVDFAGGGELFLKLARR 368
Query: 130 KYFSERRAATYVASLARALIYCHGKHVIHRDIKPENLLIGAQGELKIADFGWSVHTFNRR 189
+ F E A Y L AL +CH + HRD+KP+NLL+ A+G LK++DFG S +
Sbjct: 369 RKFPEALARRYFQQLVSALCFCHRNGIAHRDLKPQNLLLDAEGNLKVSDFGLSALPEQLK 548
Query: 190 ----RTMCGTLDYLPPEMVESVEHDAS-VDIWSLGVLCYEFLYGVPPFEAKEHSDTYRRI 244
T CGT Y PE++ + +D S D WS GV+ Y L G PF+ +RI
Sbjct: 549 NGLLHTACGTPAYTAPEILGRIGYDGSKADAWSCGVILYVLLAGHLPFDDSNIPAMCKRI 728
Query: 245 IQVDLKFPPKPIVSSAAKDLISQMLVKDSSERLPLHKLLEHPW 287
+ D +FP +S A+ LI Q+L + R+ L + + W
Sbjct: 729 TRRDYQFP--EWISKPARYLIYQLLDPNPKTRIRLENVFGNNW 851
>TC86661 similar to GP|9798599|dbj|BAB11737.1 serine/threonine protein
kinase {Arabidopsis thaliana}, partial (58%)
Length = 1840
Score = 159 bits (401), Expect = 2e-39
Identities = 94/290 (32%), Positives = 149/290 (50%), Gaps = 6/290 (2%)
Frame = +2
Query: 4 ATETQPQPHQQHHTASSEVSGSAKDQRRWILNDFDIGKPLGRGKFGHVYLAREKTSNHIV 63
A QP P + + A D + +++GK LG G F VY AR + V
Sbjct: 209 ARSKQPPPPRMSDIEQAAAPAPA-DATTALFGKYELGKLLGCGAFAKVYHARNTENGQSV 385
Query: 64 ALKVLFKSQLQQSQVEHQLRREVEIQSHLRHPHILRLYGYFYDQKRVYLILEYAPKGELY 123
A+KV+ K ++ + + ++RE+ I S LRHP+I+RL+ + ++Y ++E+A GEL+
Sbjct: 386 AVKVINKKKISATGLAGHVKREISIMSKLRHPNIVRLHEVLATKTKIYFVMEFAKGGELF 565
Query: 124 KELQKCKYFSERRAATYVASLARALIYCHGKHVIHRDIKPENLLIGAQGELKIADFGWS- 182
++ FSE + L A+ YCH V HRD+KPENLL+ +G LK++DFG S
Sbjct: 566 AKIANKGRFSEDLSRRLFQQLISAVGYCHSHGVFHRDLKPENLLLDDKGNLKVSDFGLSA 745
Query: 183 ----VHTFNRRRTMCGTLDYLPPEMVESVEHD-ASVDIWSLGVLCYEFLYGVPPFEAKEH 237
V T+CGT Y+ PE++ +D A VDIWS G++ + + G PF
Sbjct: 746 VKEQVRIDGMLHTLCGTPAYVAPEILAKRGYDGAKVDIWSCGIILFVLVAGYLPFNDPNL 925
Query: 238 SDTYRRIIQVDLKFPPKPIVSSAAKDLISQMLVKDSSERLPLHKLLEHPW 287
YR+I + K P S K +S+++ + R+ + ++L PW
Sbjct: 926 MVMYRKIYSGEFKCP--RWFSPDLKRFLSRLMDTNPETRITVDEILRDPW 1069
>TC77782 similar to GP|13448037|gb|AAK26845.1 SOS2-like protein kinase PKS6
{Arabidopsis thaliana}, partial (77%)
Length = 1236
Score = 157 bits (397), Expect = 5e-39
Identities = 85/278 (30%), Positives = 150/278 (53%), Gaps = 4/278 (1%)
Frame = +2
Query: 22 VSGSAKDQRRWILNDFDIGKPLGRGKFGHVYLAREKTSNHIVALKVLFKSQLQQSQVEHQ 81
+S + R + +++GK +G G F V LA+ + VA+K+L ++ + + + Q
Sbjct: 146 MSNKVPPRPRTRVGKYEMGKTIGEGSFAKVKLAKNVENGDYVAIKILDRNHVLKHNMMDQ 325
Query: 82 LRREVEIQSHLRHPHILRLYGYFYDQKRVYLILEYAPKGELYKELQKCKYFSERRAATYV 141
L+RE+ + HP++++++ + ++Y++LE GEL+ ++ E A +Y
Sbjct: 326 LKREISAMKIINHPNVIKIFEVMASKTKIYIVLELVNGGELFDKIATHGRLKEDEARSYF 505
Query: 142 ASLARALIYCHGKHVIHRDIKPENLLIGAQGELKIADFGWSVHTFNR---RRTMCGTLDY 198
L A+ YCH + V HRD+KPENLL+ G LK++DFG S ++ RT CGT +Y
Sbjct: 506 QQLINAVDYCHSRGVYHRDLKPENLLLDTNGVLKVSDFGLSTYSQQEDELLRTACGTPNY 685
Query: 199 LPPEMVESVEH-DASVDIWSLGVLCYEFLYGVPPFEAKEHSDTYRRIIQVDLKFPPKPIV 257
+ PE++ + +S DIWS GV+ + + G PF+ YR+I + D K P
Sbjct: 686 VAPEVLNDRGYVGSSSDIWSCGVILFVLMAGYLPFDEPNLIALYRKIGRADFKCP--SWF 859
Query: 258 SSAAKDLISQMLVKDSSERLPLHKLLEHPWIVQNAEPS 295
S AK L++ +L + R+ + ++LE W + +P+
Sbjct: 860 SPEAKKLLTSILNPNPLTRIKIPEILEDEWFRKGYKPA 973
>TC77210 similar to GP|21954717|gb|AAM83095.1 SOS2-like protein kinase
{Glycine max}, partial (88%)
Length = 1831
Score = 154 bits (388), Expect = 5e-38
Identities = 84/257 (32%), Positives = 145/257 (55%), Gaps = 6/257 (2%)
Frame = +1
Query: 37 FDIGKPLGRGKFGHVYLAREKTSNHIVALKVLFKSQLQQSQVEHQLRREVEIQSHLRHPH 96
+++G+ LG G F VY A+ + VA+KV+ K ++ + + Q++RE+ + ++HP+
Sbjct: 355 YELGRVLGHGTFAKVYHAKNLNTGKNVAMKVVGKEKVIKVGMIEQIKREISVMKMVKHPN 534
Query: 97 ILRLYGYFYDQKRVYLILEYAPKGELYKELQKCKYFSERRAATYVASLARALIYCHGKHV 156
I++L+ + ++Y+ +E GEL+ ++ K + E A Y L A+ +CH + V
Sbjct: 535 IVQLHEVMASKSKIYIAMELVRGGELFNKIVKGR-LKEDVARVYFQQLISAVDFCHSRGV 711
Query: 157 IHRDIKPENLLIGAQGELKIADFGWSVHTFNRRR-----TMCGTLDYLPPEMVESVEHD- 210
HRD+KPENLL+ G LK++DFG + + R+ T CGT Y+ PE++ +D
Sbjct: 712 YHRDLKPENLLLDEDGNLKVSDFGLCTFSEHLRQDGLLHTTCGTPAYVSPEVIAKKGYDG 891
Query: 211 ASVDIWSLGVLCYEFLYGVPPFEAKEHSDTYRRIIQVDLKFPPKPIVSSAAKDLISQMLV 270
A DIWS GV+ Y L G PF+ Y++I + D K P P S A+ LI+++L
Sbjct: 892 AKADIWSCGVILYVLLAGFLPFQDDNLVAMYKKIYRGDFKCP--PWFSLEARRLITKLLD 1065
Query: 271 KDSSERLPLHKLLEHPW 287
+ + R+ + K+++ W
Sbjct: 1066PNPNTRISISKIMDSSW 1116
>TC77939 similar to GP|14194167|gb|AAK56278.1 At2g26980/T20P8.3 {Arabidopsis
thaliana}, partial (70%)
Length = 1353
Score = 153 bits (386), Expect = 9e-38
Identities = 82/260 (31%), Positives = 135/260 (51%), Gaps = 10/260 (3%)
Frame = +1
Query: 37 FDIGKPLGRGKFGHVYLAREKTSNHIVALKVLFKSQLQQSQVEHQLRREVEIQSHLRHPH 96
+++G+ +G G F V AR + VALK+L K ++ + ++ Q++RE+ ++HP+
Sbjct: 553 YEVGRTIGEGTFAKVKFARNSETGEAVALKILDKEKVLKHKMAEQIKREIATMKLIKHPN 732
Query: 97 ILRLYGYFYDQKRVYLILEYAPKGELYKELQKCKYFSERRAATYVASLARALIYCHGKHV 156
++RLY + ++Y++LE+ GEL+ ++ E A Y L + YCH + V
Sbjct: 733 VVRLYEVMGSRTKIYIVLEFVTGGELFDKIVNHGRMGEPEARRYFQQLINVVDYCHSRGV 912
Query: 157 IHRDIKPENLLIGAQGELKIADFGWSVHTFNRR-----RTMCGTLDYLPPEMVESVEHD- 210
HRD+KPENLL+ A G LK++DFG S + R T CGT +Y+ PE++ +D
Sbjct: 913 YHRDLKPENLLLDAYGNLKVSDFGLSALSQQVRDDGLLHTTCGTPNYVAPEVLNDRGYDG 1092
Query: 211 ASVDIWSLGVLCYEFLYGVPPFEAKEHSDTYRRIIQVDLKFPPKPIVSSAAKDLIS---- 266
A+ D+WS GV+ + + G PF+ + Y++I D PP +
Sbjct: 1093ATADLWSCGVILFVLVAGYLPFDDPNLMELYKKISSADFTCPPVAFFQCKESGETNLDDV 1272
Query: 267 QMLVKDSSERLPLHKLLEHP 286
+ + KDS E K E P
Sbjct: 1273EAVFKDSEEHHVTEKKEEQP 1332
>TC77188 similar to PIR|T04862|T04862 probable serine/threonine-specific
protein kinase (EC 2.7.1.-) F28A21.110 - Arabidopsis
thaliana, partial (79%)
Length = 1881
Score = 150 bits (379), Expect = 6e-37
Identities = 86/272 (31%), Positives = 151/272 (54%), Gaps = 7/272 (2%)
Frame = +1
Query: 23 SGSAKDQRRWILNDFDIGKPLGRGKFGHVYLAREKTSNHIVALKVLFKSQLQQSQVEHQL 82
S + KD +L F++GK LG G F V+LA+ + VA+K++ K ++ +S + +
Sbjct: 148 SMNKKDNPNLLLGRFELGKLLGHGTFAKVHLAKNLKTGESVAIKIISKDKILKSGLVSHI 327
Query: 83 RREVEIQSHLRHPHILRLYGYFYDQKRVYLILEYAPKGELYKELQKCKYFSERRAATYVA 142
+RE+ I +RHP+I++L+ + ++Y ++EY GEL+ ++ K + E A Y
Sbjct: 328 KREISILRRVRHPNIVQLFEVMATKTKIYFVMEYVRGGELFNKVAKGR-LKEEVARKYFQ 504
Query: 143 SLARALIYCHGKHVIHRDIKPENLLIGAQGELKIADFGWSVHTFNRRR-----TMCGTLD 197
L A+ +CH + V HRD+KPENLL+ +G LK++DFG S + ++ T CGT
Sbjct: 505 QLICAVGFCHARGVFHRDLKPENLLLDEKGNLKVSDFGLSAVSDEIKQDGLFHTFCGTPA 684
Query: 198 YLPPEMVESVEHD-ASVDIWSLGVLCYEFLYGVPPF-EAKEHSDTYRRIIQVDLKFPPKP 255
Y+ PE++ +D A VDIWS GV+ + + G PF + Y++I + + + P
Sbjct: 685 YVAPEVLSRKGYDGAKVDIWSCGVVLFVLMAGYLPFHDPNNVMAMYKKIYKGEFRCP--R 858
Query: 256 IVSSAAKDLISQMLVKDSSERLPLHKLLEHPW 287
S L++++L R+ + +++E+ W
Sbjct: 859 WFSPELVSLLTRLLDIKPQTRISIPEIMENRW 954
>TC86123 homologue to PIR|S71770|S71770 calcium-dependent protein kinase (EC
2.7.1.-) - mung bean, complete
Length = 2448
Score = 147 bits (372), Expect = 4e-36
Identities = 85/263 (32%), Positives = 147/263 (55%), Gaps = 8/263 (3%)
Frame = +3
Query: 37 FDIGKPLGRGKFGHVYLAREKTSNHIVALKVLFKSQLQQSQVEHQLRREVEIQSHLR-HP 95
+ +G+ LG+G+FG +LA + +++ A K + K +L + +RRE++I HL H
Sbjct: 534 YTLGRKLGQGQFGTTFLAIDNSTSIEYACKSISKRKLISKEDVEDVRREIQIMHHLAGHK 713
Query: 96 HILRLYGYFYDQKRVYLILEYAPKGELYKELQKCKYFSERRAATYVASLARALIYCHGKH 155
+I+ + G + D V++++E GEL+ + + +++ER+AA + + CH
Sbjct: 714 NIVTIKGAYEDALYVHIVMELCSGGELFDRIIQRGHYTERKAAELTRIIVGVVEACHSLG 893
Query: 156 VIHRDIKPENLLIGAQGE---LKIADFGWSVHTFNRR--RTMCGTLDYLPPEMVESVEHD 210
V+HRD+KPEN L+ + + LK DFG SV + + G+ Y+ PE++ +
Sbjct: 894 VMHRDLKPENFLLVNKDDDFSLKAIDFGLSVFFKPGQVFTDVVGSPYYVAPEVLLK-HYG 1070
Query: 211 ASVDIWSLGVLCYEFLYGVPPFEAKEHSDTYRRIIQ--VDLKFPPKPIVSSAAKDLISQM 268
D+W+ GV+ Y L GVPPF A+ + +++ +D P P++S + KDLI +M
Sbjct: 1071PEADVWTAGVILYILLSGVPPFWAETQQGIFDAVLKGHIDFDSEPWPLISDSGKDLIRKM 1250
Query: 269 LVKDSSERLPLHKLLEHPWIVQN 291
L S+RL H++L HPWI +N
Sbjct: 1251LCSRPSDRLTAHEVLCHPWICEN 1319
>TC77566 similar to GP|310580|gb|AAA34017.1|| protein kinase 2 {Glycine
max}, partial (88%)
Length = 1502
Score = 147 bits (372), Expect = 4e-36
Identities = 84/260 (32%), Positives = 139/260 (53%), Gaps = 9/260 (3%)
Frame = +1
Query: 41 KPLGRGKFGHVYLAREKTSNHIVALKVLFKSQLQQSQVEHQLRREVEIQSHLRHPHILRL 100
K LG G FG LA++K + +VALK + + + +++ ++RE+ LRHP+I+R
Sbjct: 88 KELGSGNFGVARLAKDKNTGELVALKYIERGK----KIDENVQREIINHRSLRHPNIIRF 255
Query: 101 YGYFYDQKRVYLILEYAPKGELYKELQKCKYFSERRAATYVASLARALIYCHGKHVIHRD 160
+ ++LEYA GEL++ + FSE A + L + YCH + HRD
Sbjct: 256 KEVLLTPSHLAIVLEYASGGELFERICSAGRFSEDEARYFFQQLISGVSYCHSMEICHRD 435
Query: 161 IKPENLLI--GAQGELKIADFGWSVHTF--NRRRTMCGTLDYLPPEMVESVEHDASV-DI 215
+K EN L+ LKI DFG+S ++ ++ GT Y+ PE++ E+D + D+
Sbjct: 436 LKLENTLLDGNPSPRLKICDFGYSKSAILHSQPKSTVGTPAYIAPEVLSRKEYDGKIADV 615
Query: 216 WSLGVLCYEFLYGVPPFEAKEHSDTYR----RIIQVDLKFPPKPIVSSAAKDLISQMLVK 271
WS GV Y L G PFE E +R RII V P +S+ ++L+S++ V
Sbjct: 616 WSCGVTLYVMLVGAYPFEEPEDPRNFRKTIGRIIGVQYSIPDYVRISAECRNLLSRIFVA 795
Query: 272 DSSERLPLHKLLEHPWIVQN 291
D ++R+ + ++ ++PW ++N
Sbjct: 796 DPAKRVTIPEIKQNPWFLKN 855
>TC87242 homologue to PIR|S56716|S56716 protein kinase SPK-3 (EC 2.7.1.-) -
soybean, complete
Length = 1659
Score = 147 bits (372), Expect = 4e-36
Identities = 85/268 (31%), Positives = 140/268 (51%), Gaps = 9/268 (3%)
Frame = +1
Query: 33 ILNDFDIGKPLGRGKFGHVYLAREKTSNHIVALKVLFKSQLQQSQVEHQLRREVEIQSHL 92
+++ +++ K LG G FG L R K + +VA+K + + Q +++ + RE+ L
Sbjct: 466 MMDKYELVKDLGAGNFGVARLLRNKETKELVAMKYIERGQ----KIDENVAREIINHRSL 633
Query: 93 RHPHILRLYGYFYDQKRVYLILEYAPKGELYKELQKCKYFSERRAATYVASLARALIYCH 152
RHP+I+R + +++EYA GEL++ + FSE A + L + YCH
Sbjct: 634 RHPNIIRFKEVVLTPTHLAIVMEYAAGGELFERICNAGRFSEDEARYFFQQLISGVHYCH 813
Query: 153 GKHVIHRDIKPENLLIGAQ--GELKIADFGWSVHTF--NRRRTMCGTLDYLPPEMVESVE 208
+ HRD+K EN L+ LKI DFG+S + +R ++ GT Y+ PE++ E
Sbjct: 814 AMQICHRDLKLENTLLDGSPAPRLKICDFGYSKSSLLHSRPKSTVGTPAYIAPEVLSRRE 993
Query: 209 HDASV-DIWSLGVLCYEFLYGVPPFEAKEHSDTYRRIIQ----VDLKFPPKPIVSSAAKD 263
+D + D+WS GV Y L G PFE ++ +R+ IQ V K P +S K
Sbjct: 994 YDGKLADVWSCGVTLYVMLVGAYPFEDQDDPRNFRKTIQRIMAVQYKIPDYVHISQDCKH 1173
Query: 264 LISQMLVKDSSERLPLHKLLEHPWIVQN 291
L+S++ V + R+ L ++ HPW ++N
Sbjct: 1174LLSRIFVANPLRRISLKEIKNHPWFLKN 1257
>TC88799 similar to PIR|C71408|C71408 probable protein kinase - Arabidopsis
thaliana, partial (57%)
Length = 1648
Score = 145 bits (366), Expect = 2e-35
Identities = 92/270 (34%), Positives = 147/270 (54%), Gaps = 8/270 (2%)
Frame = +1
Query: 33 ILNDFDIGKPLGRGKFGHVYLAREKTSN-HIVALKVLFKSQLQQSQVEHQLRREVEIQSH 91
IL + + + LGRG F VY A N + VA+K++ KS+ + +E ++ RE++
Sbjct: 115 ILGKYQLTRLLGRGSFAKVYKAHSLLDNANTVAVKIIDKSKNVDAAMEPRIIREIDAMRR 294
Query: 92 LRH-PHILRLYGYFYDQKRVYLILEYAPKGELYKELQKCKYFSERRAATYVASLARALIY 150
L+H P+IL+++ + +++LI+E+A GEL+ L + FSE A Y L AL +
Sbjct: 295 LQHHPNILKIHEVMATKTKIHLIVEFAAGGELFSALSRRGRFSESTARFYFQQLVSALRF 474
Query: 151 CHGKHVIHRDIKPENLLIGAQGELKIADFGWSVHTFNRR----RTMCGTLDYLPPEMV-E 205
CH V HRD+KP+NLL+ A G LK++DFG S + + T CGT Y PE++
Sbjct: 475 CHRNGVAHRDLKPQNLLLDADGNLKVSDFGLSALPEHLKDGLLHTACGTPAYTAPEILRR 654
Query: 206 SVEHDAS-VDIWSLGVLCYEFLYGVPPFEAKEHSDTYRRIIQVDLKFPPKPIVSSAAKDL 264
S +D S D WS G++ Y L G PF+ ++I + D +FP VS A+ +
Sbjct: 655 SGGYDGSKADAWSCGLILYVLLAGYLPFDDLNIPAMCKKISRRDYRFP--DWVSKRARIV 828
Query: 265 ISQMLVKDSSERLPLHKLLEHPWIVQNAEP 294
I ++L + R+ L L + W ++ +P
Sbjct: 829 IYRLLDPNPETRMSLEGLFGNEWFKKSLKP 918
>TC85316 weakly similar to PIR|E84707|E84707 probable protein kinase
[imported] - Arabidopsis thaliana, partial (66%)
Length = 1528
Score = 145 bits (365), Expect = 2e-35
Identities = 83/262 (31%), Positives = 138/262 (51%), Gaps = 7/262 (2%)
Frame = +1
Query: 33 ILNDFDIGKPLGRGKFGHVYLAREKTSNHIVALKVLFKSQL-QQSQVEHQLRREVEIQSH 91
+ ++IG+ LG G VY A + VA+KV+ K +L + + RE+ I
Sbjct: 250 LFGKYEIGRLLGVGASAKVYHATNIETGKSVAVKVISKKKLINNGEFAANIEREISILRR 429
Query: 92 LRHPHILRLYGYFYDQKRVYLILEYAPKGELYKELQKCKYFSERRAATYVASLARALIYC 151
L HP+I+ L+ + ++Y ++E+A GEL++E+ K + +E A Y L A+ +C
Sbjct: 430 LHHPNIIDLFEVLASKSKIYFVVEFAEAGELFEEVAKKEKLTEDHARRYFRQLISAVKHC 609
Query: 152 HGKHVIHRDIKPENLLIGAQGELKIADFGWS-----VHTFNRRRTMCGTLDYLPPEMVES 206
H + V HRD+K +NLL+ LK+ DFG S + T+CGT Y+ PE++
Sbjct: 610 HSRGVFHRDLKLDNLLLDENDNLKVTDFGLSAVKNQIRPDGLLHTVCGTPSYVAPEILAK 789
Query: 207 VEHD-ASVDIWSLGVLCYEFLYGVPPFEAKEHSDTYRRIIQVDLKFPPKPIVSSAAKDLI 265
++ A D+WS GV+ + G PF + YR+I + +FP +S K+L+
Sbjct: 790 KGYEGAKADVWSCGVVLFTVTAGYLPFNDYNVTVLYRKIYRGQFRFP--KWMSCDLKNLL 963
Query: 266 SQMLVKDSSERLPLHKLLEHPW 287
S+ML + R+ + ++LE PW
Sbjct: 964 SRMLDTNPKTRISVDEILEDPW 1029
>TC78104 homologue to GP|19568098|gb|AAL89456.1 osmotic stress-activated
protein kinase {Nicotiana tabacum}, partial (95%)
Length = 1434
Score = 144 bits (363), Expect = 4e-35
Identities = 81/268 (30%), Positives = 137/268 (50%), Gaps = 9/268 (3%)
Frame = +3
Query: 33 ILNDFDIGKPLGRGKFGHVYLAREKTSNHIVALKVLFKSQLQQSQVEHQLRREVEIQSHL 92
++ +++ K +G G FG L R K + +VA+K + + +++ + RE+ L
Sbjct: 150 VMEKYEVVKDIGSGNFGVARLMRHKDTKQLVAMKYIERGH----KIDENVAREIINHRSL 317
Query: 93 RHPHILRLYGYFYDQKRVYLILEYAPKGELYKELQKCKYFSERRAATYVASLARALIYCH 152
RHP+I+R + +++EYA GEL+ + FSE A + L + YCH
Sbjct: 318 RHPNIIRFKEVVLTPTHLGIVMEYAAGGELFDRICSAGRFSEDEARYFFQQLISGVSYCH 497
Query: 153 GKHVIHRDIKPENLLIGAQ--GELKIADFGWSVHTF--NRRRTMCGTLDYLPPEMVESVE 208
+ HRD+K EN L+ LKI DFG+S + +R ++ GT Y+ PE++ E
Sbjct: 498 SMQICHRDLKLENTLLDGSPAPRLKICDFGYSKSSLLHSRPKSTVGTPAYIAPEVLSRRE 677
Query: 209 HDASV-DIWSLGVLCYEFLYGVPPFEAKEHSDTYR----RIIQVDLKFPPKPIVSSAAKD 263
+D + D+WS GV Y L G PFE ++ +R RI+ V K P +S +
Sbjct: 678 YDGKLADVWSCGVTLYVMLVGAYPFEDQDDPKNFRKTINRIMAVQYKIPDYVHISQECRH 857
Query: 264 LISQMLVKDSSERLPLHKLLEHPWIVQN 291
L+S++ V + R+ + ++ HPW ++N
Sbjct: 858 LLSRIFVASPARRITIKEIKSHPWFLKN 941
>TC86089 similar to PIR|S71172|S71172 protein kinase 41K (EC 2.7.1.-) -
Arabidopsis thaliana, partial (96%)
Length = 1835
Score = 144 bits (362), Expect = 5e-35
Identities = 80/264 (30%), Positives = 140/264 (52%), Gaps = 9/264 (3%)
Frame = +2
Query: 37 FDIGKPLGRGKFGHVYLAREKTSNHIVALKVLFKSQLQQSQVEHQLRREVEIQSHLRHPH 96
+D+ + +G G FG L ++K + +VA+K + + +++ ++RE+ LRHP+
Sbjct: 218 YDLVRDIGSGNFGIARLMQDKQTKELVAVKYIERGD----KIDENVKREIINHRSLRHPN 385
Query: 97 ILRLYGYFYDQKRVYLILEYAPKGELYKELQKCKYFSERRAATYVASLARALIYCHGKHV 156
I+R + +++EYA GEL++ + +FSE A + L + YCH V
Sbjct: 386 IVRFKEVILTPTHLAIVMEYASGGELFERISNAGHFSEDEARFFFQQLISGVSYCHAMQV 565
Query: 157 IHRDIKPENLLIGAQ--GELKIADFGWSVHT--FNRRRTMCGTLDYLPPEMVESVEHDAS 212
HRD+K EN L+ LKI DFG+S + ++ ++ GT Y+ PE++ E+D
Sbjct: 566 CHRDLKLENTLLDGSPTPRLKICDFGYSKSSVLHSQPKSTVGTPAYIAPEVLLRQEYDGK 745
Query: 213 V-DIWSLGVLCYEFLYGVPPF----EAKEHSDTYRRIIQVDLKFPPKPIVSSAAKDLISQ 267
+ D+WS GV Y L G PF E K+ T +R++ V P ++ + LIS+
Sbjct: 746 IADVWSCGVTLYVMLVGSYPFEDPNEPKDFRKTIQRVLSVQYSIPDNVQITPECRHLISR 925
Query: 268 MLVKDSSERLPLHKLLEHPWIVQN 291
+ V D +ER+ + ++ +H W ++N
Sbjct: 926 IFVFDPAERITMPEIWKHKWFLKN 997
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.321 0.137 0.415
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,420,757
Number of Sequences: 36976
Number of extensions: 156216
Number of successful extensions: 1674
Number of sequences better than 10.0: 541
Number of HSP's better than 10.0 without gapping: 1348
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1378
length of query: 300
length of database: 9,014,727
effective HSP length: 96
effective length of query: 204
effective length of database: 5,465,031
effective search space: 1114866324
effective search space used: 1114866324
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 58 (26.9 bits)
Medicago: description of AC138017.6


