

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC138015.15 + phase: 0 /pseudo
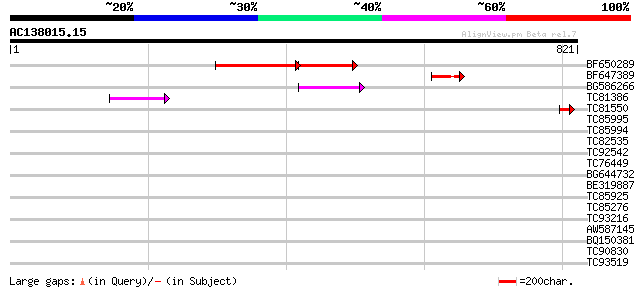
(821 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BF650289 weakly similar to GP|9049283|dbj| orf129a {Beta vulgari... 188 5e-83
BF647389 62 9e-10
BG586266 similar to GP|7267666|em RNA-directed DNA polymerase-li... 60 3e-09
TC81386 similar to GP|10140689|gb|AAG13524.1 putative non-LTR re... 46 7e-05
TC81550 similar to GP|16588669|gb|AAL26863.1 phosphoenolpyruvate... 42 7e-04
TC85995 similar to PIR|E86203|E86203 probable signal peptidase [... 33 0.34
TC85994 similar to PIR|E84708|E84708 probable signal peptidase I... 32 0.76
TC82535 similar to GP|17381261|gb|AAL36049.1 At1g61590/T25B24_6 ... 32 0.99
TC92542 homologue to PIR|T34014|T34014 hypothetical protein Y4C6... 30 2.9
TC76449 similar to GP|9293996|dbj|BAB01899.1 gene_id:MFD22.1~unk... 30 3.8
BG644732 30 4.9
BE319887 similar to PIR|T50778|T507 copper chaperone homolog CCH... 30 4.9
TC85925 weakly similar to GP|20259099|gb|AAM14265.1 putative ubi... 30 4.9
TC85276 weakly similar to GP|22294973|dbj|BAC08801. ORF_ID:tlr12... 30 4.9
TC93216 similar to PIR|T51606|T51606 probable 26S proteasome non... 30 4.9
AW587145 similar to GP|20259099|gb putative ubiquitin-conjugatin... 30 4.9
BQ150381 similar to GP|595768|gb|AA non-functional lacZ alpha pe... 29 8.4
TC90830 29 8.4
TC93519 similar to GP|10176862|dbj|BAB10068. emb|CAB80929.1~gene... 29 8.4
>BF650289 weakly similar to GP|9049283|dbj| orf129a {Beta vulgaris}, partial
(69%)
Length = 616
Score = 188 bits (478), Expect(2) = 5e-83
Identities = 100/123 (81%), Positives = 109/123 (88%)
Frame = +1
Query: 299 SKTCCAQSSQPWRSKMRYSPWILLRLQVLMVTVFVSLNALGTLLVIVSLMLY*NSLRLVS 358
+KTCCA SSQPW+SKMRYSPW LL+LQVLMVT+F+SLNALGTLLVIVSLMLY* SLRL S
Sbjct: 1 NKTCCAPSSQPWKSKMRYSPWTLLKLQVLMVTMFISLNALGTLLVIVSLMLY*ISLRLDS 180
Query: 359 CLILSTILM*LCYPKKLMIHQLRILGPLPVVQ*YIRLFPKF*QVECKES*IVLLVKINML 418
CL L T+LM*LC PKKLM+HQLRILGP VVQ*+IRLF KF*Q ECKES*IVL VKIN+L
Sbjct: 181 CLKLLTVLM*LCSPKKLMLHQLRILGPSLVVQ*FIRLFLKF*QAECKES*IVL*VKINLL 360
Query: 419 FVK 421
K
Sbjct: 361 LSK 369
Score = 139 bits (349), Expect(2) = 5e-83
Identities = 70/85 (82%), Positives = 73/85 (85%)
Frame = +3
Query: 419 FVKGRVIFDNIILSHELVKSYSRKGISPRCMVKIDLQKAYYSVE*PFINYLMLEMGFPCK 478
FVKGRVIFDNIILSHELVKSYSRKGISPRCMVKIDL KAY S E PFI +LMLE+GFP K
Sbjct: 360 FVKGRVIFDNIILSHELVKSYSRKGISPRCMVKIDLXKAYDSXEWPFIKHLMLELGFPYK 539
Query: 479 FVN*AMACLTTTSYTYNVNGDLTRP 503
FVN MA LTT SYT+N NGDLT P
Sbjct: 540 FVNWVMAXLTTASYTFNXNGDLTXP 614
>BF647389
Length = 404
Score = 62.0 bits (149), Expect = 9e-10
Identities = 35/48 (72%), Positives = 36/48 (74%)
Frame = +3
Query: 611 SLSLTSSKVRFLLGTLGSPCPPKNYMLFSVNLW*RRSFVELKIGLPIC 658
SLSL SSK FLLGTLG CPPKNYMLF+ *RRSFVELKI L C
Sbjct: 3 SLSLISSKANFLLGTLGFLCPPKNYMLFN---G*RRSFVELKIDLLSC 137
>BG586266 similar to GP|7267666|em RNA-directed DNA polymerase-like protein
{Arabidopsis thaliana}, partial (18%)
Length = 789
Score = 60.5 bits (145), Expect = 3e-09
Identities = 31/98 (31%), Positives = 54/98 (54%), Gaps = 2/98 (2%)
Frame = -3
Query: 419 FVKGRVIFDNIILSHELVKSYSRKGISPRCM--VKIDLQKAYYSVE*PFINYLMLEMGFP 476
FV GR I DN++++H+++ + G VK D+ KAY + F+ ++ +GF
Sbjct: 661 FVPGRAISDNVLITHKILHYLRQSGAKKHVSMAVKTDMTKAYDRIAWNFLREVLTRLGFH 482
Query: 477 CKFVN*AMACLTTTSYTYNVNGDLTRPFAAKKGLRQGD 514
+++ M C++T SY++ +NG +GLRQGD
Sbjct: 481 GIWISWIMECVSTVSYSFLINGGPQGRVLPSRGLRQGD 368
>TC81386 similar to GP|10140689|gb|AAG13524.1 putative non-LTR retroelement
reverse transcriptase {Oryza sativa (japonica
cultivar-group)}, partial (1%)
Length = 798
Score = 45.8 bits (107), Expect = 7e-05
Identities = 30/87 (34%), Positives = 43/87 (48%)
Frame = -1
Query: 145 KLKALKKDLKELNSTHFQGTTKKVEDARTALVDVQRQLSSDPMNLDLIEAEKVCLSSLKK 204
KLK LK+ LK N F +V++A L D+Q ++ S + L++ EK +L+
Sbjct: 306 KLKNLKEVLKVWNKNTFGNVHSQVDNAYKELDDIQVKIDSIGYSDVLMDQEKAAQLNLES 127
Query: 205 WSTIEEQIWI*KSRANWIQLGDSTQNF 231
IEE W KS+ NW GD F
Sbjct: 126 ALNIEEVFWHEKSKVNWHCEGDRNTAF 46
>TC81550 similar to GP|16588669|gb|AAL26863.1 phosphoenolpyruvate
carboxylase housekeeping isozyme pepc2 {Phaseolus
vulgaris}, partial (42%)
Length = 681
Score = 42.4 bits (98), Expect = 7e-04
Identities = 17/25 (68%), Positives = 20/25 (80%), Gaps = 2/25 (8%)
Frame = +3
Query: 796 YFDNEGYRCWQAKRWLIK--VIMYM 818
YFDNEGY CW AK WLIK +I+Y+
Sbjct: 483 YFDNEGYSCWHAKHWLIKLHIIIYV 557
>TC85995 similar to PIR|E86203|E86203 probable signal peptidase [imported] -
Arabidopsis thaliana, partial (61%)
Length = 1571
Score = 33.5 bits (75), Expect = 0.34
Identities = 23/66 (34%), Positives = 35/66 (52%)
Frame = +3
Query: 553 FMCVLLMICCCFLEVMWILSLSFLKLLACLVLLLALKLIKLRAQFIWEVYPRAFKILLSL 612
F +LL++CC L W+ FL LLL ++L+KL F + +P + + L+ L
Sbjct: 921 FSLLLLLVCC--LNRFWLNPNLFL-------LLLCIQLLKLVIVF*QKSFPSSLQSLMCL 1073
Query: 613 SLTSSK 618
L SSK
Sbjct: 1074ILRSSK 1091
>TC85994 similar to PIR|E84708|E84708 probable signal peptidase I [imported]
- Arabidopsis thaliana, partial (64%)
Length = 973
Score = 32.3 bits (72), Expect = 0.76
Identities = 23/66 (34%), Positives = 35/66 (52%)
Frame = +1
Query: 553 FMCVLLMICCCFLEVMWILSLSFLKLLACLVLLLALKLIKLRAQFIWEVYPRAFKILLSL 612
F +LL++CC L W+ FL LLL ++L+KL F + +P + + L+ L
Sbjct: 16 FSLLLLLVCC--LNRFWLNPNLFL-------LLLCIQLLKLVIVF*QKSFPSSLESLMCL 168
Query: 613 SLTSSK 618
L SSK
Sbjct: 169 IL*SSK 186
>TC82535 similar to GP|17381261|gb|AAL36049.1 At1g61590/T25B24_6
{Arabidopsis thaliana}, partial (37%)
Length = 677
Score = 32.0 bits (71), Expect = 0.99
Identities = 18/57 (31%), Positives = 32/57 (55%)
Frame = +2
Query: 555 CVLLMICCCFLEVMWILSLSFLKLLACLVLLLALKLIKLRAQFIWEVYPRAFKILLS 611
C+L++IC F V ++LSL +++CLV ++ I++ I + R +LLS
Sbjct: 260 CLLVLICLIFS*VNFVLSLRIFLVISCLVKVVLELFIRVILMIILDKV*RLNLLLLS 430
>TC92542 homologue to PIR|T34014|T34014 hypothetical protein Y4C6B.4 -
Caenorhabditis elegans, partial (1%)
Length = 516
Score = 30.4 bits (67), Expect = 2.9
Identities = 12/33 (36%), Positives = 18/33 (54%)
Frame = -2
Query: 557 LLMICCCFLEVMWILSLSFLKLLACLVLLLALK 589
+L C FL ++WI + +K L +L ALK
Sbjct: 104 ILFATCAFLNILWIFAFCAMKFLCIFILF*ALK 6
>TC76449 similar to GP|9293996|dbj|BAB01899.1 gene_id:MFD22.1~unknown
protein {Arabidopsis thaliana}, partial (24%)
Length = 1154
Score = 30.0 bits (66), Expect = 3.8
Identities = 19/44 (43%), Positives = 22/44 (49%), Gaps = 2/44 (4%)
Frame = -2
Query: 556 VLLMICCCFLEVMWILSLSFLK--LLACLVLLLALKLIKLRAQF 597
VLL CCCF FLK LL CLVL+L + + L F
Sbjct: 859 VLLCCCCCF----------FLKSVLLGCLVLVLVVAFVVLEILF 758
>BG644732
Length = 248
Score = 29.6 bits (65), Expect = 4.9
Identities = 12/29 (41%), Positives = 20/29 (68%)
Frame = +1
Query: 133 NLHTNLLTNIWFKLKALKKDLKELNSTHF 161
+L + + N+W+KL ALK L++LN+ F
Sbjct: 109 DLAKDSMKNVWYKLMALKPLLRQLNNREF 195
>BE319887 similar to PIR|T50778|T507 copper chaperone homolog CCH [imported]
- soybean, partial (20%)
Length = 587
Score = 29.6 bits (65), Expect = 4.9
Identities = 12/39 (30%), Positives = 25/39 (63%)
Frame = +3
Query: 80 HWVKFFLKEYLITVLYVWICLVLLLQRILLSYLLMY*LT 118
+W+ + + +++T+L+ I ++LLLQ L + + *LT
Sbjct: 141 NWMVYLIPYFILTILFYSILIILLLQNFLCYPICVV*LT 257
>TC85925 weakly similar to GP|20259099|gb|AAM14265.1 putative
ubiquitin-conjugating enzyme E2 {Arabidopsis thaliana},
partial (52%)
Length = 3277
Score = 29.6 bits (65), Expect = 4.9
Identities = 12/27 (44%), Positives = 17/27 (62%)
Frame = +1
Query: 187 MNLDLIEAEKVCLSSLKKWSTIEEQIW 213
+N +L E+ KVCLS L WS ++W
Sbjct: 2539 INPNLYESGKVCLSLLNTWSGTATEVW 2619
>TC85276 weakly similar to GP|22294973|dbj|BAC08801.
ORF_ID:tlr1249~hypothetical protein {Thermosynechococcus
elongatus BP-1}, partial (33%)
Length = 1315
Score = 29.6 bits (65), Expect = 4.9
Identities = 22/62 (35%), Positives = 33/62 (52%), Gaps = 2/62 (3%)
Frame = -2
Query: 534 SLGKMLLSTSTQDVKG*I*FMCVLLMICCC--FLEVMWILSLSFLKLLACLVLLLALKLI 591
++ K++LS+ST + L CCC F+ + I L+ L +L LVLLL L L+
Sbjct: 1143 TMAKLMLSSSTSSIS------TTLSSFCCCASFVVLDLIFCLALLSIL--LVLLLLLLLL 988
Query: 592 KL 593
L
Sbjct: 987 LL 982
>TC93216 similar to PIR|T51606|T51606 probable 26S proteasome non-ATPase
chain S5a [imported] - rice, partial (45%)
Length = 661
Score = 29.6 bits (65), Expect = 4.9
Identities = 16/62 (25%), Positives = 32/62 (50%)
Frame = +1
Query: 287 ISSREVLCYLSISKTCCAQSSQPWRSKMRYSPWILLRLQVLMVTVFVSLNALGTLLVIVS 346
I ++ + Y S+ + + Q W S+ S WI L + V ++T+ + TLLV+ +
Sbjct: 19 IFQKKSVLYSFKSRFRISSTFQQWCSRRHLSLWITLNICVTVITLLLDFKLNLTLLVLFA 198
Query: 347 LM 348
++
Sbjct: 199 VL 204
>AW587145 similar to GP|20259099|gb putative ubiquitin-conjugating enzyme E2
{Arabidopsis thaliana}, partial (16%)
Length = 611
Score = 29.6 bits (65), Expect = 4.9
Identities = 12/27 (44%), Positives = 17/27 (62%)
Frame = +3
Query: 187 MNLDLIEAEKVCLSSLKKWSTIEEQIW 213
+N +L E+ KVCLS L WS ++W
Sbjct: 411 INPNLYESGKVCLSLLNTWSGTATEVW 491
>BQ150381 similar to GP|595768|gb|AA non-functional lacZ alpha peptide
{unidentified cloning vector}, partial (10%)
Length = 545
Score = 28.9 bits (63), Expect = 8.4
Identities = 27/87 (31%), Positives = 42/87 (48%), Gaps = 1/87 (1%)
Frame = +2
Query: 553 FMCVLLMICCCFLEVMWILSLSFLKLLACLVLLLALKLIKLRAQFIWEVYPRAFKILLSL 612
F CVL +CC F V++I + +L C+ + L+L F +Y + +LS
Sbjct: 173 FFCVLSFLCCPFFCVVYIRPVVCHSVL-CVCV*LSL-------SFFGSLYLSFLRHMLSR 328
Query: 613 SLTSSKVRFLLGT-LGSPCPPKNYMLF 638
S+ S V F L + S C ++Y LF
Sbjct: 329 SIGSFFVVFSLSLFIASVC--RHYSLF 403
>TC90830
Length = 603
Score = 28.9 bits (63), Expect = 8.4
Identities = 16/68 (23%), Positives = 34/68 (49%), Gaps = 7/68 (10%)
Frame = -3
Query: 66 DLLLTLVGCKGSTMHWVKFFLKEYLITVLYVWICLVL-------LLQRILLSYLLMY*LT 118
DL L+ G+T+H + + I ++ +++C ++ L++ L + + Y LT
Sbjct: 307 DLSSILLHATGNTLHCTSNLVFCFFILLIILFLCSIIHILCL*FLIETFCLHFFIAYNLT 128
Query: 119 IIFFPSLI 126
+FF S +
Sbjct: 127 NLFFGSTL 104
>TC93519 similar to GP|10176862|dbj|BAB10068.
emb|CAB80929.1~gene_id:MRO11.1~similar to unknown
protein {Arabidopsis thaliana}, partial (12%)
Length = 1151
Score = 28.9 bits (63), Expect = 8.4
Identities = 18/61 (29%), Positives = 32/61 (51%), Gaps = 5/61 (8%)
Frame = -3
Query: 555 CVLLMICCCFLEVMWILSLSFLKLL--ACLV---LLLALKLIKLRAQFIWEVYPRAFKIL 609
CVL ++C C L + IL + ++LL CL+ LL + ++ + F+ + +IL
Sbjct: 987 CVLFIVCICLLNLSCILFIVCIRLLNIGCLLDMRCLLNMLIVGITLNFMMVHFMLIVRIL 808
Query: 610 L 610
L
Sbjct: 807 L 805
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.348 0.153 0.523
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 30,797,801
Number of Sequences: 36976
Number of extensions: 526262
Number of successful extensions: 6349
Number of sequences better than 10.0: 38
Number of HSP's better than 10.0 without gapping: 2777
Number of HSP's successfully gapped in prelim test: 362
Number of HSP's that attempted gapping in prelim test: 3368
Number of HSP's gapped (non-prelim): 3471
length of query: 821
length of database: 9,014,727
effective HSP length: 104
effective length of query: 717
effective length of database: 5,169,223
effective search space: 3706332891
effective search space used: 3706332891
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 14 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.8 bits)
S2: 62 (28.5 bits)
Medicago: description of AC138015.15


