

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC138012.5 + phase: 0
(182 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
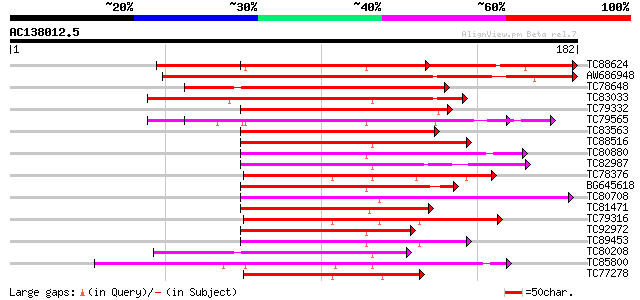
Score E
Sequences producing significant alignments: (bits) Value
TC88624 similar to PIR|S51529|S51529 SPF1 protein - sweet potato... 135 7e-33
AW686948 homologue to PIR|S51529|S515 SPF1 protein - sweet potat... 119 7e-28
TC78648 similar to PIR|S51529|S51529 SPF1 protein - sweet potato... 105 1e-23
TC83033 similar to PIR|S51529|S51529 SPF1 protein - sweet potato... 94 2e-20
TC79332 similar to PIR|JC6203|JC6203 SP8 binding protein homolog... 91 2e-19
TC79565 similar to SP|Q9ZQ70|WRK3_ARATH Probable WRKY transcript... 89 7e-19
TC83563 similar to PIR|T52092|T52092 DNA-binding protein WRKY2 [... 89 1e-18
TC88516 similar to GP|14530681|dbj|BAB61053. WRKY DNA-binding pr... 75 1e-14
TC80880 similar to SP|Q9ZQ70|WRK3_ARATH Probable WRKY transcript... 75 1e-14
TC82987 similar to GP|15991740|gb|AAL13047.1 WRKY transcription ... 73 5e-14
TC78376 similar to SP|Q9SK33|WR60_ARATH Probable WRKY transcript... 70 4e-13
BG645618 similar to GP|18158619|gb WRKY-like drought-induced pro... 70 4e-13
TC80708 weakly similar to GP|14530685|dbj|BAB61055. WRKY DNA-bin... 70 5e-13
TC81471 similar to PIR|A84913|A84913 probable WRKY-type DNA bind... 69 8e-13
TC79316 similar to GP|11493822|gb|AAG35658.1 transcription facto... 68 2e-12
TC92972 similar to GP|9187622|emb|CAB97004.1 WRKY DNA binding pr... 68 2e-12
TC89453 similar to GP|9187622|emb|CAB97004.1 WRKY DNA binding pr... 65 1e-11
TC80208 similar to GP|17064158|gb|AAL35286.1 WRKY transcription ... 65 2e-11
TC85800 similar to GP|11320830|dbj|BAB18313. putative WRKY DNA b... 65 2e-11
TC77278 similar to GP|11493822|gb|AAG35658.1 transcription facto... 63 6e-11
>TC88624 similar to PIR|S51529|S51529 SPF1 protein - sweet potato, partial
(50%)
Length = 1496
Score = 135 bits (341), Expect = 7e-33
Identities = 69/140 (49%), Positives = 94/140 (66%), Gaps = 5/140 (3%)
Frame = +2
Query: 48 DNNRNEFQSDYSNGRQQNKSSVRRMNH--DDGYNWKKYEEKVAKGSENQRSYYKCTWPNC 105
+N N FQSDYSN +Q + + ++ DDGYNW+KY +K KGSEN RSYYKCT+PNC
Sbjct: 680 NNTNNGFQSDYSNYQQPQQQPTQTLSRRSDDGYNWRKYGQKQVKGSENPRSYYKCTYPNC 859
Query: 106 FVKKKVERTIDGEVIETLYKGTHNHWKPTSSMKRNSSSEYLYSLLPSETGSIDLQDQSF- 164
KKKVER+I+G+V E +YKGTHNH KP + + +SSS ++P + ++ DQS+
Sbjct: 860 PTKKKVERSIEGQVTEIVYKGTHNHPKPQCTRRNSSSSSNALVVVPVNPIN-EIHDQSYA 1036
Query: 165 --GSEQLDSDEEPTKSSVSI 182
G+ Q+DS P SS+SI
Sbjct: 1037SHGNGQMDSAATPENSSISI 1096
Score = 76.3 bits (186), Expect = 6e-15
Identities = 35/62 (56%), Positives = 43/62 (68%), Gaps = 1/62 (1%)
Frame = +2
Query: 75 DDGYNWKKYEEKVAKGSENQRSYYKCTWPNCFVKKKVER-TIDGEVIETLYKGTHNHWKP 133
DDGY W+KY +KV KG+ N RSYYKCT PNC V+K VER + D + T Y+G HNH P
Sbjct: 1298 DDGYRWRKYGQKVVKGNPNPRSYYKCTHPNCPVRKHVERASHDLRAVITTYEGKHNHDVP 1477
Query: 134 TS 135
+
Sbjct: 1478 AA 1483
>AW686948 homologue to PIR|S51529|S515 SPF1 protein - sweet potato, partial
(16%)
Length = 565
Score = 119 bits (298), Expect = 7e-28
Identities = 63/134 (47%), Positives = 86/134 (64%), Gaps = 1/134 (0%)
Frame = +1
Query: 50 NRNEFQSDYSNGRQQNKSSVRRMNHDDGYNWKKYEEKVAKGSENQRSYYKCTWPNCFVKK 109
N++ QS+Y+N + Q + + DDGYNW+KY +K KGSEN RSYYKCT+PNC KK
Sbjct: 100 NKSGSQSNYNNYQSQPQVQILSRRSDDGYNWRKYGQKQVKGSENPRSYYKCTYPNCPTKK 279
Query: 110 KVERTIDGEVIETLYKGTHNHWKPTSSMKRNSSSEYLYSLLPSETGSIDLQDQSFGSE-Q 168
KVER +DG++ E +YKG+HNH KP ++ KRN++S SL + FG+E Q
Sbjct: 280 KVERGLDGQITEIVYKGSHNHPKPVAN-KRNTNSMSSSSLSHANPP----PSNHFGNEIQ 444
Query: 169 LDSDEEPTKSSVSI 182
+D P SS+SI
Sbjct: 445 MDLVATPENSSISI 486
>TC78648 similar to PIR|S51529|S51529 SPF1 protein - sweet potato, partial
(18%)
Length = 990
Score = 105 bits (262), Expect = 1e-23
Identities = 48/85 (56%), Positives = 61/85 (71%)
Frame = +1
Query: 57 DYSNGRQQNKSSVRRMNHDDGYNWKKYEEKVAKGSENQRSYYKCTWPNCFVKKKVERTID 116
+Y+N Q +R +DGYNW+KY +K KGSEN RSYYKCT PNC +KKKVER +D
Sbjct: 721 NYNNNASQFVREQKRS--EDGYNWRKYGQKQVKGSENPRSYYKCTNPNCSMKKKVERDLD 894
Query: 117 GEVIETLYKGTHNHWKPTSSMKRNS 141
G++ E +YKGTHNH KP S+ + NS
Sbjct: 895 GQITEIVYKGTHNHPKPQSNRRNNS 969
>TC83033 similar to PIR|S51529|S51529 SPF1 protein - sweet potato, partial
(17%)
Length = 900
Score = 94.4 bits (233), Expect = 2e-20
Identities = 47/110 (42%), Positives = 69/110 (62%), Gaps = 7/110 (6%)
Frame = +3
Query: 45 DGVDNNRNEFQSDYSNGRQQNKSSV------RRMNHDDGYNWKKYEEKVAKGSENQRSYY 98
+G + + S N N +S +R + +DGY W+KY EK KGSEN RSYY
Sbjct: 474 EGTTKHEAQTNSSVPNSSYFNNTSASMSIREQRKSEEDGYKWRKYGEKQVKGSENPRSYY 653
Query: 99 KCTWPNCFVKKKVERTI-DGEVIETLYKGTHNHWKPTSSMKRNSSSEYLY 147
KCT P C + KKVER++ DG++ + +YKG+HNH KP + KR ++S+Y++
Sbjct: 654 KCTNPICSMXKKVERSLDDGQITQIVYKGSHNHPKPQCT-KRRANSQYIH 800
>TC79332 similar to PIR|JC6203|JC6203 SP8 binding protein homolog -
cucumber, partial (49%)
Length = 1486
Score = 91.3 bits (225), Expect = 2e-19
Identities = 43/70 (61%), Positives = 50/70 (71%), Gaps = 2/70 (2%)
Frame = +2
Query: 75 DDGYNWKKYEEKVAKGSENQRSYYKCTWPNCFVKKKVERTIDGEVIETLYKGTHNHWKPT 134
DDGYNW+KY +K KGSE RSYYKCT NC VKKKVER DG + E +YKG HNH KP
Sbjct: 725 DDGYNWRKYGQKQVKGSEYPRSYYKCTHLNCPVKKKVERAPDGHITEIIYKGQHNHEKPQ 904
Query: 135 SS--MKRNSS 142
+ +K N+S
Sbjct: 905 PNRRVKENNS 934
Score = 32.0 bits (71), Expect = 0.14
Identities = 11/17 (64%), Positives = 14/17 (81%)
Frame = +3
Query: 75 DDGYNWKKYEEKVAKGS 91
DDGY W+KY +KV KG+
Sbjct: 1272 DDGYRWRKYGQKVVKGN 1322
>TC79565 similar to SP|Q9ZQ70|WRK3_ARATH Probable WRKY transcription factor
3 (WRKY DNA-binding protein 3). [Mouse-ear cress],
partial (35%)
Length = 1602
Score = 89.4 bits (220), Expect = 7e-19
Identities = 52/124 (41%), Positives = 72/124 (57%), Gaps = 5/124 (4%)
Frame = +2
Query: 57 DYSNGRQQN--KSSVRRMN--HDDGYNWKKYEEKVAKGSENQRSYYKCTWPNCFVKKKVE 112
+YS+ +Q KSS ++ +DDGYNW+KY +K KG E RSYYKCT P+C V KKVE
Sbjct: 608 EYSSNSEQRLQKSSFVNVDKANDDGYNWRKYGQKQVKGCEFPRSYYKCTHPSCLVTKKVE 787
Query: 113 R-TIDGEVIETLYKGTHNHWKPTSSMKRNSSSEYLYSLLPSETGSIDLQDQSFGSEQLDS 171
R +DG V +YKG H H +P S N +S L +G+ D +++ G + +
Sbjct: 788 RDPVDGHVTAIIYKGEHIHQRPRPSKLTNDNSSVQQVL----SGTSDSEEE--GDHETEV 949
Query: 172 DEEP 175
D EP
Sbjct: 950 DYEP 961
Score = 75.1 bits (183), Expect = 1e-14
Identities = 45/142 (31%), Positives = 69/142 (47%), Gaps = 25/142 (17%)
Frame = +2
Query: 45 DGVDNNRNEFQSDYSNGRQQNKSSVRRMNH-----------------------DDGYNWK 81
D + +E + DY G ++ K+ + +N +DGY W+
Sbjct: 914 DSEEEGDHETEVDYEPGLKRRKTEAKLLNPALSHRTVSKPKIIVQTTSDVDLLEDGYRWR 1093
Query: 82 KYEEKVAKGSENQRSYYKCTWPNCFVKKKVER-TIDGEVIETLYKGTHNHWKPTS-SMKR 139
KY +KV KG+ RSYYKCT P C V+K VER + D + + T Y+G HNH P + +
Sbjct: 1094 KYGQKVVKGNPYPRSYYKCTTPGCNVRKHVERVSTDPKAVLTTYEGKHNHDVPAAKTNSH 1273
Query: 140 NSSSEYLYSLLPSETGSIDLQD 161
N +S S L S+ ++Q+
Sbjct: 1274 NLASNNSASQLKSQNAIPEMQN 1339
>TC83563 similar to PIR|T52092|T52092 DNA-binding protein WRKY2 [imported] -
common tobacco, partial (21%)
Length = 838
Score = 88.6 bits (218), Expect = 1e-18
Identities = 38/64 (59%), Positives = 46/64 (71%)
Frame = +1
Query: 75 DDGYNWKKYEEKVAKGSENQRSYYKCTWPNCFVKKKVERTIDGEVIETLYKGTHNHWKPT 134
DDGYNW+KY +K KGSE RSYYKCT PNC VKKKVER++ G + +YKG HNH P
Sbjct: 646 DDGYNWRKYGQKQVKGSEFPRSYYKCTHPNCPVKKKVERSLAGHITAIIYKGEHNHLLPN 825
Query: 135 SSMK 138
+ +
Sbjct: 826 PNKR 837
>TC88516 similar to GP|14530681|dbj|BAB61053. WRKY DNA-binding protein
{Nicotiana tabacum}, partial (17%)
Length = 1085
Score = 75.5 bits (184), Expect = 1e-14
Identities = 36/75 (48%), Positives = 46/75 (61%), Gaps = 1/75 (1%)
Frame = +2
Query: 75 DDGYNWKKYEEKVAKGSENQRSYYKCTWPNCFVKKKVERTI-DGEVIETLYKGTHNHWKP 133
DDG+ W+KY +KV KG+ N RSYYKCT P C V+K VER D + + T Y+G HNH P
Sbjct: 284 DDGFRWRKYGQKVVKGNPNARSYYKCTAPGCNVRKHVERAAHDIKAVITTYEGKHNHDVP 463
Query: 134 TSSMKRNSSSEYLYS 148
+ SE +S
Sbjct: 464 AARGSAGLQSEQKFS 508
>TC80880 similar to SP|Q9ZQ70|WRK3_ARATH Probable WRKY transcription factor
3 (WRKY DNA-binding protein 3). [Mouse-ear cress],
partial (24%)
Length = 907
Score = 75.1 bits (183), Expect = 1e-14
Identities = 40/93 (43%), Positives = 55/93 (59%), Gaps = 1/93 (1%)
Frame = +2
Query: 75 DDGYNWKKYEEKVAKGSENQRSYYKCTWPNCFVKKKVER-TIDGEVIETLYKGTHNHWKP 133
DDGY W+KY +KV KG+ RSYYKCT P C V+K VER + D + + T Y+G HNH P
Sbjct: 236 DDGYRWRKYGQKVVKGNPYPRSYYKCTTPGCNVRKHVERASTDPKAVITTYEGKHNHDVP 415
Query: 134 TSSMKRNSSSEYLYSLLPSETGSIDLQDQSFGS 166
+ ++ + S L S+ + + SFGS
Sbjct: 416 AAKTNSHTIANNNASQLKSQ--NTISEKTSFGS 508
>TC82987 similar to GP|15991740|gb|AAL13047.1 WRKY transcription factor 71
{Arabidopsis thaliana}, partial (33%)
Length = 975
Score = 73.2 bits (178), Expect = 5e-14
Identities = 39/94 (41%), Positives = 56/94 (59%), Gaps = 1/94 (1%)
Frame = +1
Query: 75 DDGYNWKKYEEKVAKGSENQRSYYKCTWPNCFVKKKVERTI-DGEVIETLYKGTHNHWKP 133
+DGY W+KY +K K S RSYY+CT C VKK+VER+ D ++ T Y+G HNH P
Sbjct: 679 EDGYRWRKYGQKAVKNSPYPRSYYRCTSQKCIVKKRVERSYQDPSIVMTTYEGQHNHHCP 858
Query: 134 TSSMKRNSSSEYLYSLLPSETGSIDLQDQSFGSE 167
++++ N+S S+L S + S+ S G E
Sbjct: 859 -ATLRGNAS-----SMLLSSSPSLFASSTSMGLE 942
>TC78376 similar to SP|Q9SK33|WR60_ARATH Probable WRKY transcription factor
60 (WRKY DNA-binding protein 60). [Mouse-ear cress],
partial (38%)
Length = 1121
Score = 70.5 bits (171), Expect = 4e-13
Identities = 36/87 (41%), Positives = 58/87 (66%), Gaps = 6/87 (6%)
Frame = +1
Query: 76 DGYNWKKYEEKVAKGSENQRSYYKCTW-PNCFVKKKVERTI-DGEVIETLYKGTHN--HW 131
DGY W+KY +KV + + + R+Y+KC++ P C VKKKV+R++ D V+ T Y+G HN H
Sbjct: 595 DGYQWRKYGQKVTRDNPSPRAYFKCSFAPGCPVKKKVQRSVEDQNVLVTTYEGEHNHAHH 774
Query: 132 KPTSSMKRNSSSEY--LYSLLPSETGS 156
+P S+ ++ SE Y+L+P+ + S
Sbjct: 775 QPEMSLTSSNQSETTPTYNLVPASSSS 855
>BG645618 similar to GP|18158619|gb WRKY-like drought-induced protein {Retama
raetam}, partial (37%)
Length = 747
Score = 70.5 bits (171), Expect = 4e-13
Identities = 35/71 (49%), Positives = 47/71 (65%), Gaps = 1/71 (1%)
Frame = -1
Query: 75 DDGYNWKKYEEKVAKGSENQRSYYKCTWPNCFVKKKVER-TIDGEVIETLYKGTHNHWKP 133
DDGY W+KY +KV +G+ N RSYYKCT C V+K VER + D + + T Y+G HNH P
Sbjct: 384 DDGYRWRKYGQKVVRGNPNPRSYYKCTNAGCPVRKHVERASHDPKAVITTYEGKHNHDVP 205
Query: 134 TSSMKRNSSSE 144
+ R+SS +
Sbjct: 204 AA---RSSSHD 181
Score = 31.2 bits (69), Expect = 0.24
Identities = 12/26 (46%), Positives = 19/26 (72%)
Frame = -1
Query: 118 EVIETLYKGTHNHWKPTSSMKRNSSS 143
++IE +YKGTH+H KP S + ++ S
Sbjct: 747 QIIEIIYKGTHDHPKPQPSNRYSAGS 670
>TC80708 weakly similar to GP|14530685|dbj|BAB61055. WRKY DNA-binding
protein {Nicotiana tabacum}, partial (20%)
Length = 864
Score = 70.1 bits (170), Expect = 5e-13
Identities = 35/108 (32%), Positives = 60/108 (55%), Gaps = 1/108 (0%)
Frame = +2
Query: 75 DDGYNWKKYEEKVAKGSENQRSYYKCTWPNCFVKKKVERTIDG-EVIETLYKGTHNHWKP 133
+DGY W+KY +K+ KG+ N R+YY+C+ P C VKK VE++ + T Y+G H+H P
Sbjct: 191 NDGYRWRKYGQKMVKGNTNPRNYYRCSSPGCPVKKHVEKSSQNTTTVITTYEGQHDHAPP 370
Query: 134 TSSMKRNSSSEYLYSLLPSETGSIDLQDQSFGSEQLDSDEEPTKSSVS 181
T ++++ L + + + L+ + EQ + D + S+S
Sbjct: 371 TGRGVLDNTAVKLTPIRATILPNQSLESELNEEEQRNEDPSTKEDSIS 514
>TC81471 similar to PIR|A84913|A84913 probable WRKY-type DNA binding protein
[imported] - Arabidopsis thaliana, partial (43%)
Length = 1274
Score = 69.3 bits (168), Expect = 8e-13
Identities = 31/63 (49%), Positives = 41/63 (64%), Gaps = 1/63 (1%)
Frame = +2
Query: 75 DDGYNWKKYEEKVAKGSENQRSYYKCTWPNCFVKKKVERT-IDGEVIETLYKGTHNHWKP 133
+DGY W+KY +K K S RSYY+CT +C VKK+VER+ D ++ T Y+G H H P
Sbjct: 557 EDGYRWRKYGQKAVKNSPFPRSYYRCTTASCNVKKRVERSYTDPSIVVTTYEGQHTHPSP 736
Query: 134 TSS 136
T S
Sbjct: 737 TMS 745
>TC79316 similar to GP|11493822|gb|AAG35658.1 transcription factor WRKY4
{Petroselinum crispum}, partial (49%)
Length = 1282
Score = 67.8 bits (164), Expect = 2e-12
Identities = 35/90 (38%), Positives = 55/90 (60%), Gaps = 7/90 (7%)
Frame = +3
Query: 76 DGYNWKKYEEKVAKGSENQRSYYKCTW-PNCFVKKKVERTIDG-EVIETLYKGTHNH--- 130
DGY W+KY +KV + + + R+Y+KC++ P+C VKKKV+R++D V+ Y+G HNH
Sbjct: 603 DGYQWRKYGQKVTRDNPSPRAYFKCSFAPSCTVKKKVQRSVDDPSVVVATYEGEHNHPHP 782
Query: 131 --WKPTSSMKRNSSSEYLYSLLPSETGSID 158
+PT++ RN S S T ++D
Sbjct: 783 SQTEPTNNRCRNQISPVAPSSSAPSTVTLD 872
>TC92972 similar to GP|9187622|emb|CAB97004.1 WRKY DNA binding protein
{Solanum tuberosum}, partial (46%)
Length = 672
Score = 67.8 bits (164), Expect = 2e-12
Identities = 29/57 (50%), Positives = 42/57 (72%), Gaps = 1/57 (1%)
Frame = +2
Query: 75 DDGYNWKKYEEKVAKGSENQRSYYKCTWPNCFVKKKVER-TIDGEVIETLYKGTHNH 130
DDG+ W+KY EK+ K ++ RSYYKCT+ C VKK+++R + D +V+ET Y+G H H
Sbjct: 230 DDGFRWRKYGEKMVKDNKFPRSYYKCTYQGCKVKKQIQRHSKDEQVVETSYEGMHIH 400
>TC89453 similar to GP|9187622|emb|CAB97004.1 WRKY DNA binding protein
{Solanum tuberosum}, partial (56%)
Length = 701
Score = 65.5 bits (158), Expect = 1e-11
Identities = 33/77 (42%), Positives = 46/77 (58%), Gaps = 3/77 (3%)
Frame = +1
Query: 75 DDGYNWKKYEEKVAKGSENQRSYYKCTWPNCFVKKKVER-TIDGEVIETLYKGTHNH--W 131
DDGY W+KY +K K ++ RSYY+CT C VKK+V+R T D V+ T Y+G H H
Sbjct: 463 DDGYRWRKYGQKAVKNNKFPRSYYRCTHQGCNVKKQVQRLTKDEGVVVTTYEGVHTHPIE 642
Query: 132 KPTSSMKRNSSSEYLYS 148
K T + + S +Y+
Sbjct: 643 KTTDNFEHILSXMQIYT 693
>TC80208 similar to GP|17064158|gb|AAL35286.1 WRKY transcription factor 28
{Arabidopsis thaliana}, partial (31%)
Length = 1294
Score = 64.7 bits (156), Expect = 2e-11
Identities = 34/84 (40%), Positives = 46/84 (54%), Gaps = 1/84 (1%)
Frame = +1
Query: 47 VDNNRNEFQSDYSNGRQQNKSSVRRMNHDDGYNWKKYEEKVAKGSENQRSYYKCTWPNCF 106
VD + E + KS V ++ +DGY W+KY +K K S RSYY+CT C
Sbjct: 529 VDKKKGEKKQKEPRFAFMTKSEVDQL--EDGYRWRKYGQKAVKNSPYPRSYYRCTTQKCT 702
Query: 107 VKKKVERTI-DGEVIETLYKGTHN 129
VKK+VER+ D + T Y+G HN
Sbjct: 703 VKKRVERSYEDPTTVITTYEGQHN 774
>TC85800 similar to GP|11320830|dbj|BAB18313. putative WRKY DNA binding
protein {Oryza sativa (japonica cultivar-group)},
partial (37%)
Length = 2028
Score = 64.7 bits (156), Expect = 2e-11
Identities = 49/149 (32%), Positives = 74/149 (48%), Gaps = 15/149 (10%)
Frame = +1
Query: 28 RHEIGTLRVEDDVAIEIDGVDNNRNEFQSDYSNGRQQNKS---------SVRRMNH---- 74
R+ G ED E G N+++ + SN Q + SVR +
Sbjct: 841 RNSNGKTGREDSPESETQGWGPNKSQKILNSSNVADQANTEATMRKARVSVRARSEASMI 1020
Query: 75 DDGYNWKKYEEKVAKGSENQRSYYKCTWP-NCFVKKKVERTI-DGEVIETLYKGTHNHWK 132
DG W+KY +K+AKG+ R+YY+CT C V+K+V+R D ++ T Y+GTHNH
Sbjct: 1021SDGCQWRKYGQKMAKGNPCPRAYYRCTMAVGCPVRKQVQRCAEDKTILVTTYEGTHNHPL 1200
Query: 133 PTSSMKRNSSSEYLYSLLPSETGSIDLQD 161
P ++M S++ S+L S GS+ D
Sbjct: 1201PPAAMAMASTTSAAASMLLS--GSMSSAD 1281
>TC77278 similar to GP|11493822|gb|AAG35658.1 transcription factor WRKY4
{Petroselinum crispum}, partial (40%)
Length = 1604
Score = 63.2 bits (152), Expect = 6e-11
Identities = 27/60 (45%), Positives = 45/60 (75%), Gaps = 2/60 (3%)
Frame = +2
Query: 76 DGYNWKKYEEKVAKGSENQRSYYKCTW-PNCFVKKKVERTIDGE-VIETLYKGTHNHWKP 133
DGY+W+KY +KV + + R+Y+KC++ P+C VKKKV+R++D + ++ Y+G HNH +P
Sbjct: 692 DGYHWRKYGQKVTRDNPCPRAYFKCSFAPSCPVKKKVQRSVDDQSMLVATYEGEHNHPQP 871
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.311 0.129 0.368
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,702,919
Number of Sequences: 36976
Number of extensions: 76484
Number of successful extensions: 479
Number of sequences better than 10.0: 89
Number of HSP's better than 10.0 without gapping: 450
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 453
length of query: 182
length of database: 9,014,727
effective HSP length: 90
effective length of query: 92
effective length of database: 5,686,887
effective search space: 523193604
effective search space used: 523193604
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 55 (25.8 bits)
Medicago: description of AC138012.5


