

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC138011.4 + phase: 0
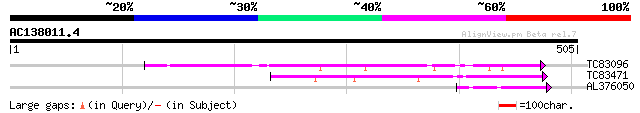
(505 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC83096 weakly similar to PIR|T45912|T45912 hypothetical protein... 158 5e-39
TC83471 similar to PIR|T45912|T45912 hypothetical protein F5K20.... 135 4e-32
AL376050 similar to GP|10178015|dbj Na+/H+ antiporter-like prote... 42 7e-04
>TC83096 weakly similar to PIR|T45912|T45912 hypothetical protein F5K20.20 -
Arabidopsis thaliana, partial (21%)
Length = 1227
Score = 158 bits (399), Expect = 5e-39
Identities = 119/376 (31%), Positives = 182/376 (47%), Gaps = 19/376 (5%)
Frame = +2
Query: 121 IYKPRKRFEQNKLKTIQKLRLDAELRILTCVHNARQATGVISLVESFNAT-RISPIHVFA 179
IYKP ++ + + +LRIL C+H +I+ +ES AT + S I ++
Sbjct: 11 IYKPSRQRRSGNPPPLTDTQ--EKLRILACIHGTGNIPSLINFIESVRATNKSSKIKLYV 184
Query: 180 LYLIELKGRTGALVAAHMDKPSNQPGAQNLTRSQVEQESINNTFEALGEAYDAVRVETLN 239
+ L EL + +++ + S P + +++ F A G+ V V L
Sbjct: 185 MQLTELTDSSSSILMVRSSRKSGFPFINRFQKGTMQE-----AFRACGQV-GQVTVHHLT 346
Query: 240 VVSSYSTIHEDIYNSANEKRTSMIILPFHKHLSSLG--ALETTSVAYKDINQNVMQTSPC 297
+SS STIHEDI + A EK +MIILPFHK +E ++++NQ V+Q++PC
Sbjct: 347 SISSLSTIHEDICHIAEEKGVAMIILPFHKRWRGEDEETIEDIGQRWREVNQRVLQSAPC 526
Query: 298 SVGIFVDRNLGSITKMNF--------RICMIFIGGPDDHEALAVAWRMVGHPGTRLSLVR 349
SV + V+R +G + ++C+IF+GGPDD + L + RM HP RLS+VR
Sbjct: 527 SVAVLVNRGVGRRYEQRVETSATPGKKVCIIFVGGPDDRKVLELGSRMAEHPAIRLSVVR 706
Query: 350 MLLFDEAAEVNIISHYEVQGILSVVMDN--EKQKDLDEEYVNSFRLKAVNNNDSISYSEV 407
L +E + Y S DN E +K+LDE +N F+ K + ++ Y E
Sbjct: 707 FNLHNEGTFRDQEHSYNTSTSAS---DNNMENEKELDEVALNEFKTKWLG---AVEYIE- 865
Query: 408 DVHSSEDIPTILNDLDKFG----YDLYIVGQGHH--RNVRAFSSLLEWCDCPELGVIGDM 461
D I N++ G Y+L IVG+GH + + ELG IGD+
Sbjct: 866 -----NDTVNIANEVLAIGRVKEYELVIVGKGHQLLNSTGMIDIKDSQLEHAELGPIGDL 1030
Query: 462 LASNNFSSGSSVLVVQ 477
L S+ SSVLV+Q
Sbjct: 1031LTSSAQGITSSVLVIQ 1078
>TC83471 similar to PIR|T45912|T45912 hypothetical protein F5K20.20 -
Arabidopsis thaliana, partial (18%)
Length = 983
Score = 135 bits (340), Expect = 4e-32
Identities = 90/272 (33%), Positives = 140/272 (51%), Gaps = 25/272 (9%)
Frame = +3
Query: 233 VRVETLNVVSSYSTIHEDIYNSANEKRTSMIILPFHKHL----------SSLGALETTSV 282
V V + +SS ST+HEDI ++A EKR +MIILPFHKH + LE
Sbjct: 15 VIVRSTTAISSLSTMHEDICHAAEEKRVTMIILPFHKHWRMEVDDENDKEAHEVLENAGH 194
Query: 283 AYKDINQNVMQTSPCSVGIFVDR-------NLGSITKMNFRICMIFIGGPDDHEALAVAW 335
++ +NQ V++ +PCSV + VDR NLGS ++ RIC++F GGPDD EAL +
Sbjct: 195 GWRGVNQRVLKNAPCSVAVLVDRGYGLGLKNLGSDGRVAQRICIVFFGGPDDREALELGK 374
Query: 336 RMVGHPGTRLSLVRMLLFDEAAEVNII-------SHYEVQGILSVVMDNEKQKDLDEEYV 388
+MV HP +++VR + +E + N + S E ++ +K++ LDE +
Sbjct: 375 KMVEHPAVVVTVVRFVEQNELSGNNFVLRQSPGKSTEENYSFSIAKINRQKEQVLDENAM 554
Query: 389 NSFRLKAVNNNDSISYSEVDVHSSEDIPTILNDLDKFGYDLYIVGQGHHRNVRAFSSLLE 448
FR K +++ Y ++ S + ++ + YDL +VG+G +
Sbjct: 555 EEFRSKC---GETVKY--IEKGSGNVVEEVIALGESADYDLIVVGKGRFPSTMVAELAER 719
Query: 449 WCDCPELGVIGDMLASN-NFSSGSSVLVVQQY 479
+ ELG IGD+L S+ SSV V+QQ+
Sbjct: 720 EAEHAELGPIGDILTSSMGHKMASSVFVIQQH 815
>AL376050 similar to GP|10178015|dbj Na+/H+ antiporter-like protein
{Arabidopsis thaliana}, partial (3%)
Length = 521
Score = 41.6 bits (96), Expect = 7e-04
Identities = 30/85 (35%), Positives = 46/85 (53%), Gaps = 1/85 (1%)
Frame = +2
Query: 399 NDSISYSEVDVHSSEDIPTILNDLDKFGY-DLYIVGQGHHRNVRAFSSLLEWCDCPELGV 457
+DS+ Y E V +D + + KF +L++VG R + LE +CPELG
Sbjct: 2 DDSVIYEEKIV---KDAAETVASIRKFNSCNLFLVGL---RPTGELACALERRECPELGP 163
Query: 458 IGDMLASNNFSSGSSVLVVQQYGYG 482
+G +L S + + +SVLV+QQY G
Sbjct: 164 VGGLLISQDCPTTASVLVMQQYHNG 238
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.320 0.135 0.388
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 13,766,928
Number of Sequences: 36976
Number of extensions: 168571
Number of successful extensions: 866
Number of sequences better than 10.0: 6
Number of HSP's better than 10.0 without gapping: 854
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 859
length of query: 505
length of database: 9,014,727
effective HSP length: 100
effective length of query: 405
effective length of database: 5,317,127
effective search space: 2153436435
effective search space used: 2153436435
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 60 (27.7 bits)
Medicago: description of AC138011.4


