

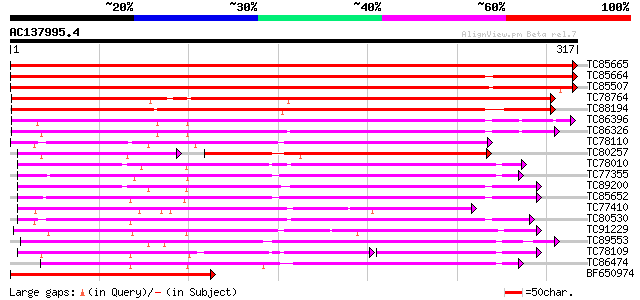
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137995.4 - phase: 0
(317 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC85665 similar to GP|18157333|dbj|BAB83762. 1-aminocyclopropane... 646 0.0
TC85664 1-aminocyclopropanecarboxylic acid oxidase [Medicago tru... 565 e-162
TC85507 homologue to SP|P31239|ACCO_PEA 1-aminocyclopropane-1-ca... 491 e-139
TC78764 similar to PIR|C96802|C96802 hypothetical protein F2P24.... 303 4e-83
TC88194 similar to PIR|F84578|F84578 1-aminocyclopropane-1-carbo... 285 1e-77
TC86396 similar to GP|10178137|dbj|BAB11549. leucoanthocyanidin ... 207 6e-54
TC86326 similar to GP|6016680|gb|AAF01507.1| putative leucoantho... 202 1e-52
TC78110 similar to PIR|S44261|S44261 SRG1 protein - Arabidopsis ... 181 3e-46
TC80257 similar to PIR|S44261|S44261 SRG1 protein - Arabidopsis ... 137 3e-43
TC78010 similar to PIR|S44261|S44261 SRG1 protein - Arabidopsis ... 171 3e-43
TC77355 weakly similar to PIR|T05552|T05552 SRG1 protein-related... 170 6e-43
TC89200 similar to PIR|S44261|S44261 SRG1 protein - Arabidopsis ... 169 1e-42
TC85652 similar to PIR|S44261|S44261 SRG1 protein - Arabidopsis ... 165 2e-41
TC77410 similar to GP|21392365|gb|AAM48289.1 flavanone 3 beta-hy... 165 2e-41
TC80530 weakly similar to PIR|S44261|S44261 SRG1 protein - Arabi... 163 7e-41
TC91229 similar to GP|9294689|dbj|BAB03055.1 gene_id:MHC9.10~sim... 160 8e-40
TC89553 weakly similar to SP|P24397|HY6H_HYONI Hyoscyamine 6-dio... 158 2e-39
TC78109 similar to PIR|S44261|S44261 SRG1 protein - Arabidopsis ... 109 3e-39
TC86474 weakly similar to GP|6016680|gb|AAF01507.1| putative leu... 153 7e-38
BF650974 similar to SP|P31239|ACCO 1-aminocyclopropane-1-carboxy... 149 1e-36
>TC85665 similar to GP|18157333|dbj|BAB83762.
1-aminocyclopropane-1-carboxylic acid oxidase {Phaseolus
lunatus}, partial (97%)
Length = 1284
Score = 646 bits (1667), Expect = 0.0
Identities = 317/317 (100%), Positives = 317/317 (100%)
Frame = +2
Query: 1 MMNFPIISLEKLNGVERKDTMEKIKDACENWGFFELVNHGIPHDLMDTLERLTKEHYRKC 60
MMNFPIISLEKLNGVERKDTMEKIKDACENWGFFELVNHGIPHDLMDTLERLTKEHYRKC
Sbjct: 47 MMNFPIISLEKLNGVERKDTMEKIKDACENWGFFELVNHGIPHDLMDTLERLTKEHYRKC 226
Query: 61 MEHRFKEVISSKGLDVVQTEVKDMDWESTFHVRHLPESNISEIPDLSDEYRKVMKEFSLR 120
MEHRFKEVISSKGLDVVQTEVKDMDWESTFHVRHLPESNISEIPDLSDEYRKVMKEFSLR
Sbjct: 227 MEHRFKEVISSKGLDVVQTEVKDMDWESTFHVRHLPESNISEIPDLSDEYRKVMKEFSLR 406
Query: 121 LEKLAEELLDLLCENLGLEKGYLKKAFYGSRGPTFGTKVANYPQCPNPELVKGLRAHTDA 180
LEKLAEELLDLLCENLGLEKGYLKKAFYGSRGPTFGTKVANYPQCPNPELVKGLRAHTDA
Sbjct: 407 LEKLAEELLDLLCENLGLEKGYLKKAFYGSRGPTFGTKVANYPQCPNPELVKGLRAHTDA 586
Query: 181 GGIILLFQDDKVSGLQLLKDGQWVDVPPMRHSIVVNLGDQLEVITNGKYKSVEHRVIAQT 240
GGIILLFQDDKVSGLQLLKDGQWVDVPPMRHSIVVNLGDQLEVITNGKYKSVEHRVIAQT
Sbjct: 587 GGIILLFQDDKVSGLQLLKDGQWVDVPPMRHSIVVNLGDQLEVITNGKYKSVEHRVIAQT 766
Query: 241 NGTRMSIASFYNPGSDAVIYPAPELLEKQTEEKHNVYPKFVFEEYMKIYAALKFHAKEPR 300
NGTRMSIASFYNPGSDAVIYPAPELLEKQTEEKHNVYPKFVFEEYMKIYAALKFHAKEPR
Sbjct: 767 NGTRMSIASFYNPGSDAVIYPAPELLEKQTEEKHNVYPKFVFEEYMKIYAALKFHAKEPR 946
Query: 301 FEALKESNVNLGPIAIV 317
FEALKESNVNLGPIAIV
Sbjct: 947 FEALKESNVNLGPIAIV 997
>TC85664 1-aminocyclopropanecarboxylic acid oxidase [Medicago truncatula]
Length = 1832
Score = 565 bits (1457), Expect = e-162
Identities = 274/317 (86%), Positives = 292/317 (91%)
Frame = +1
Query: 1 MMNFPIISLEKLNGVERKDTMEKIKDACENWGFFELVNHGIPHDLMDTLERLTKEHYRKC 60
M NFPII+LE LNG ERK TMEKIKDACENWGFFELVNHGIPHDLMDT+ERLTKEHYRKC
Sbjct: 637 MDNFPIINLENLNGDERKATMEKIKDACENWGFFELVNHGIPHDLMDTVERLTKEHYRKC 816
Query: 61 MEHRFKEVISSKGLDVVQTEVKDMDWESTFHVRHLPESNISEIPDLSDEYRKVMKEFSLR 120
ME RFKE++SSKGL+ VQTEVKDMDWESTFH+RHLPESNISEIPDLSDEYRK MKEF+L+
Sbjct: 817 MEQRFKELVSSKGLEAVQTEVKDMDWESTFHLRHLPESNISEIPDLSDEYRKSMKEFALK 996
Query: 121 LEKLAEELLDLLCENLGLEKGYLKKAFYGSRGPTFGTKVANYPQCPNPELVKGLRAHTDA 180
LE LAEELLDLLCENLGLEKGYLKKA YGS+GPTFGTKVANYP CP P+LVKGLRAHTDA
Sbjct: 997 LETLAEELLDLLCENLGLEKGYLKKALYGSKGPTFGTKVANYPPCPKPDLVKGLRAHTDA 1176
Query: 181 GGIILLFQDDKVSGLQLLKDGQWVDVPPMRHSIVVNLGDQLEVITNGKYKSVEHRVIAQT 240
GGIILLFQDDKVSGLQLLKDG WVDVPPM HSIV+NLGDQLEVITNGKYKSVEHRV+AQT
Sbjct: 1177GGIILLFQDDKVSGLQLLKDGNWVDVPPMHHSIVINLGDQLEVITNGKYKSVEHRVVAQT 1356
Query: 241 NGTRMSIASFYNPGSDAVIYPAPELLEKQTEEKHNVYPKFVFEEYMKIYAALKFHAKEPR 300
+GTRMSIASFYNPGSDAVIYPAP L+ EE + +YPKFVFE+YM +YA LKF AKEPR
Sbjct: 1357DGTRMSIASFYNPGSDAVIYPAPTLI----EENNEIYPKFVFEDYMNLYARLKFQAKEPR 1524
Query: 301 FEALKESNVNLGPIAIV 317
FEA KESNVNLGPIA V
Sbjct: 1525FEAFKESNVNLGPIATV 1575
>TC85507 homologue to SP|P31239|ACCO_PEA 1-aminocyclopropane-1-carboxylate
oxidase (EC 1.-.-.-) (ACC oxidase) (Ethylene-forming
enzyme) (EFE), complete
Length = 1342
Score = 491 bits (1264), Expect = e-139
Identities = 233/319 (73%), Positives = 274/319 (85%), Gaps = 2/319 (0%)
Frame = +1
Query: 1 MMNFPIISLEKLNGVERKDTMEKIKDACENWGFFELVNHGIPHDLMDTLERLTKEHYRKC 60
M NFP++ + KLN ERK TME IKDACENWGFFE VNH I +LMD +E+LTKEHY+KC
Sbjct: 97 MENFPVVDMGKLNTEERKATMEMIKDACENWGFFECVNHSISIELMDKVEKLTKEHYKKC 276
Query: 61 MEHRFKEVISSKGLDVVQTEVKDMDWESTFHVRHLPESNISEIPDLSDEYRKVMKEFSLR 120
ME RFKE+++SKGL+ VQ+E+ D+DWESTF +RHLP SNISEIPDL ++YRK MKEF+ +
Sbjct: 277 MEQRFKEMVASKGLECVQSEINDLDWESTFFLRHLPSSNISEIPDLDEDYRKTMKEFAEK 456
Query: 121 LEKLAEELLDLLCENLGLEKGYLKKAFYGSRGPTFGTKVANYPQCPNPELVKGLRAHTDA 180
LEKLAEELLDLLCENLGLEKGYLKK FYGS+GP FGTKV+NYP CP P+L+KGLRAHTDA
Sbjct: 457 LEKLAEELLDLLCENLGLEKGYLKKVFYGSKGPNFGTKVSNYPPCPKPDLIKGLRAHTDA 636
Query: 181 GGIILLFQDDKVSGLQLLKDGQWVDVPPMRHSIVVNLGDQLEVITNGKYKSVEHRVIAQT 240
GGIILLFQDDKVSGLQLLKD QW+DVPPM HSIV+NLGDQLEVITNGKYKSV HRVIAQT
Sbjct: 637 GGIILLFQDDKVSGLQLLKDDQWIDVPPMPHSIVINLGDQLEVITNGKYKSVMHRVIAQT 816
Query: 241 NGTRMSIASFYNPGSDAVIYPAPELLEKQTEEKHNVYPKFVFEEYMKIYAALKFHAKEPR 300
+G RMSIASFYNPG+DAVI PA LL++ +E +YPKF+F++YMK+Y LKF AKEPR
Sbjct: 817 DGARMSIASFYNPGNDAVISPASTLLKE--DETSEIYPKFIFDDYMKLYMGLKFQAKEPR 990
Query: 301 FEALKE--SNVNLGPIAIV 317
FEA+ + S+V +GP+ +
Sbjct: 991 FEAMMKAMSSVEVGPVVTI 1047
>TC78764 similar to PIR|C96802|C96802 hypothetical protein F2P24.4
[imported] - Arabidopsis thaliana, partial (88%)
Length = 1276
Score = 303 bits (777), Expect = 4e-83
Identities = 154/311 (49%), Positives = 212/311 (67%), Gaps = 7/311 (2%)
Frame = +1
Query: 2 MNFPIISLEKLNGVERKDTMEKIKDACENWGFFELVNHGIPHDLMDTLERLTKEHYRKCM 61
M P+I KLNG ER T+ +I + CE WGFF+L+NHGI +L++ +++++ E Y+
Sbjct: 67 MAVPVIDFSKLNGEERAKTLAQIANGCEEWGFFQLINHGISEELLERVKKVSSEFYKLER 246
Query: 62 EHRFKEVISSKGLDVV-----QTEVKDMDWESTFHVRHLPESNISEIPDLSDEYRKVMKE 116
E FK + K L+ + +++++DWE V L + N E P+ + +R+ M E
Sbjct: 247 EENFKNSKTVKLLNDIAEKKSSEKLENVDWED---VITLLDDN--EWPENTPSFRETMSE 411
Query: 117 FSLRLEKLAEELLDLLCENLGLEKGYLKKAFYGSRGPT--FGTKVANYPQCPNPELVKGL 174
+ L+KLA L +++ ENLGL KGY+KKA G FGTKV++YP CP+PELV GL
Sbjct: 412 YRSELKKLAVSLTEVMDENLGLPKGYIKKALNDGEGDNAFFGTKVSHYPPCPHPELVNGL 591
Query: 175 RAHTDAGGIILLFQDDKVSGLQLLKDGQWVDVPPMRHSIVVNLGDQLEVITNGKYKSVEH 234
RAHTDAGG+ILLFQDDKV GLQ+LKDG+W+DV P+ ++IV+N GDQ+EV++NG+ KS H
Sbjct: 592 RAHTDAGGVILLFQDDKVGGLQMLKDGEWLDVQPLPNAIVINTGDQIEVLSNGRNKSCWH 771
Query: 235 RVIAQTNGTRMSIASFYNPGSDAVIYPAPELLEKQTEEKHNVYPKFVFEEYMKIYAALKF 294
RV+ T GTR +IASFYNP A I PAP+L EK ++ + YPKFVF +YM +YA KF
Sbjct: 772 RVLNFTEGTRRTIASFYNPPLKATISPAPQLAEKDNQQVDDTYPKFVFGDYMSVYAEQKF 951
Query: 295 HAKEPRFEALK 305
KEPRF A+K
Sbjct: 952 LPKEPRFRAVK 984
>TC88194 similar to PIR|F84578|F84578 1-aminocyclopropane-1-carboxylate
oxidase [imported] - Arabidopsis thaliana, partial (91%)
Length = 1018
Score = 285 bits (730), Expect = 1e-77
Identities = 144/308 (46%), Positives = 207/308 (66%), Gaps = 4/308 (1%)
Frame = +2
Query: 2 MNFPIISLEKLNGVERKDTMEKIKDACENWGFFELVNHGIPHDLMDTLERLTKEHYRKCM 61
M P+I LNG +R +TM + +AC+ WG F + NH I LM+ ++++ +Y + +
Sbjct: 2 MAIPVIDFSTLNGDKRGETMALLHEACQKWGCFLIENHDIEGKLMEKVKKVINSYYEENL 181
Query: 62 EHRFKEVISSKGLDVVQTEVKDMDWESTFHVRHLPESNISEIPDLSDEYRKVMKEFSLRL 121
+ F + +K L+ + D+DWES+F + H P SNI +IP+LS++ + M E+ +L
Sbjct: 182 KESFYQSEIAKRLEKKENTC-DVDWESSFFIWHRPTSNIRKIPNLSEDLCQTMDEYIDKL 358
Query: 122 EKLAEELLDLLCENLGLEKGYLKKAFYGSR--GPTFGTKVANYPQCPNPELVKGLRAHTD 179
++AE L ++ ENLGLEK Y+KKAF G+ GP GTKVA YP+CP PELV+GLR HTD
Sbjct: 359 VQVAETLSQMMSENLGLEKDYIKKAFSGNNNNGPAMGTKVAKYPECPYPELVRGLREHTD 538
Query: 180 AGGIILLFQDDKVSGLQLLKDGQWVDVPPMR-HSIVVNLGDQLEVITNGKYKSVEHRVIA 238
AGGIILL QDDKV GL+ KDG+W+++PP + ++I VN GDQ+EV++NG YKS HRV+
Sbjct: 539 AGGIILLLQDDKVPGLEFFKDGKWIEIPPSKNNAIFVNTGDQIEVLSNGLYKSAVHRVMP 718
Query: 239 QTNGTRMSIASFYNPGSDAVIYPAPELLEKQTEEKHNVYP-KFVFEEYMKIYAALKFHAK 297
NG+R+SIASFYNP +A+I PAP+LL YP + + +Y+++Y KF K
Sbjct: 719 DKNGSRLSIASFYNPVGEAIISPAPKLL----------YPSNYCYGDYLELYGKTKFGDK 868
Query: 298 EPRFEALK 305
PRFE++K
Sbjct: 869 GPRFESIK 892
>TC86396 similar to GP|10178137|dbj|BAB11549. leucoanthocyanidin
dioxygenase-like protein {Arabidopsis thaliana}, partial
(82%)
Length = 1326
Score = 207 bits (526), Expect = 6e-54
Identities = 116/325 (35%), Positives = 189/325 (57%), Gaps = 10/325 (3%)
Frame = +2
Query: 2 MNFPIISLEKLNG---VERKDTMEKIKDACENWGFFELVNHGIPHDLMDTLERLTKEHYR 58
+N P+I LE L+ V R+ ++++ +AC WGFF++VNHGI H+LM++ + + +E +
Sbjct: 248 INIPVIDLEHLSSEDPVLRETVLKRVSEACREWGFFQVVNHGISHELMESAKEVWREFFN 427
Query: 59 KCMEHRFKEVISSKGLDVVQTEV-----KDMDWESTFHVRHLPES--NISEIPDLSDEYR 111
+E + + S + + + +DW F + +P S N ++ P R
Sbjct: 428 LPLEVKEEFANSPSTYEGYGSRLGVKKGAILDWSDYFFLHSMPPSLRNQAKWPATPSSLR 607
Query: 112 KVMKEFSLRLEKLAEELLDLLCENLGLEKGYLKKAFYGSRGPTFGTKVANYPQCPNPELV 171
K++ E+ + KL +L+L+ NLGL++ YL AF G +V YP+CP P+L
Sbjct: 608 KIIAEYGEEVVKLGGRMLELMSTNLGLKEDYLMNAFGGENELGACLRVNFYPKCPQPDLT 787
Query: 172 KGLRAHTDAGGIILLFQDDKVSGLQLLKDGQWVDVPPMRHSIVVNLGDQLEVITNGKYKS 231
GL H+D GG+ +L DD VSGLQ+ K W+ V P+ ++ ++N+GDQ++V++N YKS
Sbjct: 788 LGLSPHSDPGGMTILLPDDFVSGLQVRKGNDWITVKPVPNAFIINIGDQIQVLSNAIYKS 967
Query: 232 VEHRVIAQTNGTRMSIASFYNPGSDAVIYPAPELLEKQTEEKHNVYPKFVFEEYMKIYAA 291
VEHRVI + R+S+A FYNP SD +I PA EL+ T+E+ +Y ++EY +++
Sbjct: 968 VEHRVIVNSIKDRVSLAMFYNPKSDLLIEPAKELV---TKERPALYSAMTYDEY-RLFIR 1135
Query: 292 LKFHAKEPRFEALKESNVNLGPIAI 316
+K + + E+L S V L PI I
Sbjct: 1136MKGPCGKTQVESL-ASQV*LVPIII 1207
>TC86326 similar to GP|6016680|gb|AAF01507.1| putative leucoanthocyanidin
dioxygenase {Arabidopsis thaliana}, partial (81%)
Length = 1686
Score = 202 bits (515), Expect = 1e-52
Identities = 115/316 (36%), Positives = 179/316 (56%), Gaps = 10/316 (3%)
Frame = +2
Query: 2 MNFPIISLEKLNGVE---RKDTMEKIKDACENWGFFELVNHGIPHDLMDTLERLTKEHYR 58
+N PII L LNG + +++I DAC +WGFF++VNHG+ DLMD ++ +
Sbjct: 392 INIPIIDLGGLNGDDLDVHASILKQISDACRDWGFFQIVNHGVSPDLMDKARETWRQFFH 571
Query: 59 KCMEHRFKEVISSKGLDVVQTEV-----KDMDWESTFHVRHLPES--NISEIPDLSDEYR 111
ME + + S + + + +DW + + +LP S + ++ P R
Sbjct: 572 LPMEAKQQYANSPTTYEGYGSRLGVEKGAILDWSDYYFLHYLPVSVKDCNKWPASPQSCR 751
Query: 112 KVMKEFSLRLEKLAEELLDLLCENLGLEKGYLKKAFYGSRGPTFGTKVANYPQCPNPELV 171
+V E+ L KL+ L+ L NLGLE+ L+ AF G +V YP+CP PEL
Sbjct: 752 EVFDEYGKELVKLSGRLMKALSLNLGLEEKILQNAFGGEEIGAC-MRVNFYPKCPRPELT 928
Query: 172 KGLRAHTDAGGIILLFQDDKVSGLQLLKDGQWVDVPPMRHSIVVNLGDQLEVITNGKYKS 231
GL +H+D GG+ +L DD+V+GLQ+ K W+ V P RH +VN+GDQ++V++N YKS
Sbjct: 929 LGLSSHSDPGGMTMLLPDDQVAGLQVRKFDNWITVNPARHGFIVNIGDQIQVLSNATYKS 1108
Query: 232 VEHRVIAQTNGTRMSIASFYNPGSDAVIYPAPELLEKQTEEKHNVYPKFVFEEYMKIYAA 291
VEHRVI ++ R+S+A FYNP SD I P +L+ T E+ +YP F+EY +++
Sbjct: 1109VEHRVIVNSDQERLSLAFFYNPRSDIPIEPLKQLI---TPERPALYPAMTFDEY-RLFIR 1276
Query: 292 LKFHAKEPRFEALKES 307
++ + + E++K S
Sbjct: 1277MRGPCGKSQVESMKSS 1324
>TC78110 similar to PIR|S44261|S44261 SRG1 protein - Arabidopsis thaliana,
partial (53%)
Length = 1303
Score = 181 bits (459), Expect = 3e-46
Identities = 99/286 (34%), Positives = 160/286 (55%), Gaps = 16/286 (5%)
Frame = +2
Query: 1 MMNFPIISLEKL-------NGVERKDTMEKIKDACENWGFFELVNHGIPHDLMDTLERLT 53
++ P+I KL G+E ++K+ AC+ WGFF+L+NHG+ L++ ++
Sbjct: 200 LLELPVIDFSKLFSQDLTIKGLE----LDKLHSACKEWGFFQLINHGVSTSLVENVKMGA 367
Query: 54 KEHYRKCMEHRFKEVISSKGLDV-------VQTEVKDMDWESTFHVRHLPESNISE--IP 104
KE Y +E + K S K DV V +E + +DW F + LP P
Sbjct: 368 KEFYNLPIEEKKK--FSQKEGDVEGYGQAFVMSEEQKLDWADMFFMITLPSHMRKPHLFP 541
Query: 105 DLSDEYRKVMKEFSLRLEKLAEELLDLLCENLGLEKGYLKKAFYGSRGPTFGTKVANYPQ 164
L +R ++ +S L+KLA +++D + L ++ +++ F T T++ YP
Sbjct: 542 KLPLPFRDDLETYSAELKKLAIQIIDFMANALKVDAKEIRELFGEG---TQSTRINYYPP 712
Query: 165 CPNPELVKGLRAHTDAGGIILLFQDDKVSGLQLLKDGQWVDVPPMRHSIVVNLGDQLEVI 224
CP PELV GL +H+D GG+ +L Q +++ GLQ+ KDG W+ V P+ ++ ++NLGD LE+I
Sbjct: 713 CPQPELVIGLNSHSDGGGLTILLQGNEMDGLQIKKDGFWIPVKPLPNAFIINLGDMLEII 892
Query: 225 TNGKYKSVEHRVIAQTNGTRMSIASFYNPGSDAVIYPAPELLEKQT 270
TNG Y S+EHR R+SIA+FY+P S ++ P+P L+ +T
Sbjct: 893 TNGIYPSIEHRATVNLKKERLSIATFYSPSSAVILRPSPTLVTPKT 1030
>TC80257 similar to PIR|S44261|S44261 SRG1 protein - Arabidopsis thaliana,
partial (50%)
Length = 1376
Score = 137 bits (345), Expect(2) = 3e-43
Identities = 64/163 (39%), Positives = 104/163 (63%), Gaps = 3/163 (1%)
Frame = +2
Query: 110 YRKVMKEFSLRLEKLAEELLDLLCENLGLEKGYLKKAFYGSRGPTFGTKVAN---YPQCP 166
+R+ ++ +S+ LEKLA ++++L+ L + + + F GT++ YP CP
Sbjct: 605 FRENLEVYSVELEKLAIKVIELMANALAINPKEMTELF------NIGTQMVRVNYYPPCP 766
Query: 167 NPELVKGLRAHTDAGGIILLFQDDKVSGLQLLKDGQWVDVPPMRHSIVVNLGDQLEVITN 226
PE V GL++H+DAGG+ +L Q + GLQ+ KDGQW+ V P+ ++ +VN+GD LE+ITN
Sbjct: 767 QPERVIGLKSHSDAGGLTILLQTSDIDGLQIRKDGQWIPVKPLPNAFIVNIGDMLEIITN 946
Query: 227 GKYKSVEHRVIAQTNGTRMSIASFYNPGSDAVIYPAPELLEKQ 269
G Y S+EHR + R+S+A+FY P A++ PAP L+ ++
Sbjct: 947 GIYPSIEHRATVNSEKERISVAAFYGPNMQAMLAPAPSLVTQE 1075
Score = 55.5 bits (132), Expect(2) = 3e-43
Identities = 30/100 (30%), Positives = 53/100 (53%), Gaps = 8/100 (8%)
Frame = +1
Query: 5 PIISLEKLNGVE---RKDTMEKIKDACENWGFFELVNHGIPHDLMDTLERLTKEHYRKCM 61
P+I L +L + ++ +EK+ AC+ WGFF+LVNHGI L++ +++ K + M
Sbjct: 259 PVIDLAELLSQDINLKRHELEKLHYACKEWGFFQLVNHGISTSLVEDMKKGAKTLFELSM 438
Query: 62 EHR-----FKEVISSKGLDVVQTEVKDMDWESTFHVRHLP 96
E + + + G V +E + ++W TF + LP
Sbjct: 439 EEKKNLWQIEGEMEGFGQSFVLSEEQKLEWADTFFLSTLP 558
>TC78010 similar to PIR|S44261|S44261 SRG1 protein - Arabidopsis thaliana,
partial (47%)
Length = 1321
Score = 171 bits (433), Expect = 3e-43
Identities = 91/294 (30%), Positives = 166/294 (55%), Gaps = 9/294 (3%)
Frame = +3
Query: 5 PIISLEKLNGVERKDTMEKIKDACENWGFFELVNHGIPHDLMDTLERLTKEHYRKCMEHR 64
P+I L KL E + ++K+ AC+ WGFF+L+NHG+ L++ ++L ++ + +E
Sbjct: 222 PVIDLSKLLSEEDETELQKLDHACKEWGFFQLINHGVNPLLVENFKKLVQDFFNLPVEE- 398
Query: 65 FKEVISSK-------GLDVVQTEVKDMDWESTFHVRHLPE--SNISEIPDLSDEYRKVMK 115
K+++S K G V +E ++W FH+ P N P + +R+ ++
Sbjct: 399 -KKILSQKPGNIEGFGQLFVVSEDHKLEWADLFHIITHPSYMRNPQLFPSIPQPFRESLE 575
Query: 116 EFSLRLEKLAEELLDLLCENLGLEKGYLKKAFYGSRGPTFGTKVANYPQCPNPELVKGLR 175
+SL L+KL +++ + + L ++K L + F+ G + ++ YP CP P+ V GL
Sbjct: 576 MYSLVLKKLCVMIIEFMSKALKIQKNELLE-FFEEGGQSM--RMNYYPPCPQPDKVIGLN 746
Query: 176 AHTDAGGIILLFQDDKVSGLQLLKDGQWVDVPPMRHSIVVNLGDQLEVITNGKYKSVEHR 235
H+D + +L Q +++ GLQ+ KDG W+ + P+ ++ V+N+GD LE++TNG Y+S+EHR
Sbjct: 747 PHSDGTALTILLQLNEIEGLQIKKDGMWIPIKPLTNAFVINIGDMLEIMTNGIYRSIEHR 926
Query: 236 VIAQTNGTRMSIASFYNPGSDAVIYPAPELLEKQTEEKHNVYPKFVFEEYMKIY 289
+ R+SIA+F++ +A++ P P L+ +T V+ E++ K Y
Sbjct: 927 ATINSEKERISIATFHSARLNAILAPVPSLITPKTPA---VFNDISVEDFFKGY 1079
>TC77355 weakly similar to PIR|T05552|T05552 SRG1 protein-related protein
F24A6.150 - Arabidopsis thaliana, partial (44%)
Length = 1359
Score = 170 bits (431), Expect = 6e-43
Identities = 96/290 (33%), Positives = 158/290 (54%), Gaps = 7/290 (2%)
Frame = +1
Query: 5 PIISLEKLNGVERKDTMEKIKDACENWGFFELVNHGIPHDLMDTLERLTKEHYRKCMEHR 64
P+I +L+ ++ + K++ AC+ WGFF++VNHG+ DL+ + + E + ME +
Sbjct: 202 PVIDFTQLSNGNMEELL-KLEIACKEWGFFQMVNHGVEKDLIQRMRDASNEFFELPMEEK 378
Query: 65 FKEV-----ISSKGLDVVQTEVKDMDWESTFHVRHLPES--NISEIPDLSDEYRKVMKEF 117
K I G + V +E + +DW + P+ + P S ++++++ +
Sbjct: 379 EKYAMLPNDIQGYGQNYVVSEEQTLDWSDVLVLIIYPDRYRKLQFWPKTSHGFKEIIEAY 558
Query: 118 SLRLEKLAEELLDLLCENLGLEKGYLKKAFYGSRGPTFGTKVANYPQCPNPELVKGLRAH 177
S + ++ EELL L +GLEK L + + G +V YP C NPE V GL H
Sbjct: 559 SSEVRRVGEELLSFLSIIMGLEKHALAELH---KELIQGLRVNYYPPCNNPEQVLGLSPH 729
Query: 178 TDAGGIILLFQDDKVSGLQLLKDGQWVDVPPMRHSIVVNLGDQLEVITNGKYKSVEHRVI 237
+D I LL QDD V GL++ G WV V P+ ++V+N+GD +E+++NGKYKSVEHRV+
Sbjct: 730 SDTTTITLLIQDDDVLGLEIRNKGNWVPVKPISDALVINVGDVIEILSNGKYKSVEHRVM 909
Query: 238 AQTNGTRMSIASFYNPGSDAVIYPAPELLEKQTEEKHNVYPKFVFEEYMK 287
N R+S ASF P D + P +++ Q + +Y K + +Y++
Sbjct: 910 TNQNKRRISYASFLFPRDDVEVEPFDHMIDAQNPK---MYQKVKYGDYLR 1050
>TC89200 similar to PIR|S44261|S44261 SRG1 protein - Arabidopsis thaliana,
partial (47%)
Length = 1323
Score = 169 bits (428), Expect = 1e-42
Identities = 94/303 (31%), Positives = 163/303 (53%), Gaps = 10/303 (3%)
Frame = +3
Query: 5 PIISLEKLNGVERKDTMEKIKDACENWGFFELVNHGIPHDLMDTLERLTKEHYRKCMEHR 64
PII L KL E ++K AC+ WGFF+L+NHG+ L++ ++ +E + E
Sbjct: 285 PIIDLNKLLSEEDGTELQKFDLACKEWGFFQLINHGVDLSLVENFKKHVQEFFNLSAEE- 461
Query: 65 FKEVISSKGLDV-------VQTEVKDMDWESTFHVRHLPE--SNISEIPDLSDEYRKVMK 115
K++ K D+ V +E ++W F++ LP N + P + +R ++
Sbjct: 462 -KKIFGQKPGDMEGFGQMFVVSEEHKLEWADLFYIVTLPTYIRNPNLFPSIPQPFRDNLE 638
Query: 116 EFSLRLEKLAEELLDLLCENLGLEKGYLKKAFY-GSRGPTFGTKVANYPQCPNPELVKGL 174
+++ ++KL +++ + + L ++ L F GS+ ++ YP CP PE V GL
Sbjct: 639 MYAIEIKKLCVTIVEFMAKALKIQPNELLDFFEEGSQA----MRMNYYPPCPQPEQVIGL 806
Query: 175 RAHTDAGGIILLFQDDKVSGLQLLKDGQWVDVPPMRHSIVVNLGDQLEVITNGKYKSVEH 234
H+D G + +L Q +++ GLQ+ KDG W+ + P+ ++ V+N+GD LE++TNG Y+S+EH
Sbjct: 807 NPHSDVGALTILLQLNEIEGLQVRKDGMWIPIKPLSNAFVINIGDMLEIMTNGIYRSIEH 986
Query: 235 RVIAQTNGTRMSIASFYNPGSDAVIYPAPELLEKQTEEKHNVYPKFVFEEYMKIYAALKF 294
R R+SIA+F + +A++ PAP L+ T E V+ E++ K Y + K
Sbjct: 987 RATINAEKERISIATFNSGRLNAILCPAPSLV---TPESPAVFNNVGVEDFFKAYFSRKL 1157
Query: 295 HAK 297
K
Sbjct: 1158EGK 1166
>TC85652 similar to PIR|S44261|S44261 SRG1 protein - Arabidopsis thaliana,
partial (50%)
Length = 1489
Score = 165 bits (417), Expect = 2e-41
Identities = 93/300 (31%), Positives = 160/300 (53%), Gaps = 7/300 (2%)
Frame = +1
Query: 5 PIISLEKLNGVERKDTMEKIKDACENWGFFELVNHGIPHDLMDTLERLTKEHYRKCMEHR 64
P+I L KL G + + +EK+ AC+ WGFF+++NHG+ L++ ++ KE +E +
Sbjct: 403 PVIDLGKLLGEDATE-LEKLDLACKEWGFFQIINHGVNTSLIEKVKIGIKEFLSLPVEEK 579
Query: 65 FK-----EVISSKGLDVVQTEVKDMDWESTFHVRHLP--ESNISEIPDLSDEYRKVMKEF 117
K + G V ++ + ++W F + LP E N P + +R ++ +
Sbjct: 580 KKFWQTPNDMEGFGQMFVVSDDQKLEWADLFLITTLPLDERNPRLFPSIFQPFRDNLEIY 759
Query: 118 SLRLEKLAEELLDLLCENLGLEKGYLKKAFYGSRGPTFGTKVANYPQCPNPELVKGLRAH 177
++KL ++ + + L +E + + F T + YP CP PE V GL H
Sbjct: 760 CSEVQKLCFTIISQMEKALKIESNEVTELF---NHITQAMRWNLYPPCPQPENVIGLNPH 930
Query: 178 TDAGGIILLFQDDKVSGLQLLKDGQWVDVPPMRHSIVVNLGDQLEVITNGKYKSVEHRVI 237
+D G + +L Q +++ GLQ+ KDGQW+ V P+ ++ V+N+GD LE++TNG Y+S+EHR
Sbjct: 931 SDVGALTILLQANEIEGLQIRKDGQWIPVQPLPNAFVINIGDMLEIVTNGIYRSIEHRAT 1110
Query: 238 AQTNGTRMSIASFYNPGSDAVIYPAPELLEKQTEEKHNVYPKFVFEEYMKIYAALKFHAK 297
+ R+SIA+F+ P + ++ P P L+ T E+ + EY+K Y + K K
Sbjct: 1111VNSEKERISIAAFHRPHVNTILSPRPSLV---TPERPASFKSIAVGEYLKAYFSRKLEGK 1281
>TC77410 similar to GP|21392365|gb|AAM48289.1 flavanone 3 beta-hydroxylase
{Solanum tuberosum}, partial (98%)
Length = 1471
Score = 165 bits (417), Expect = 2e-41
Identities = 97/268 (36%), Positives = 153/268 (56%), Gaps = 11/268 (4%)
Frame = +1
Query: 5 PIISLEKLN--GVERKDTMEKIKDACENWGFFELVNHGIPHDLMDTLERLTKEHYRKCME 62
PIISL+ ++ G R + KI +ACENWG F++V+HG+ L+ + R K + E
Sbjct: 175 PIISLDGIDDAGGRRAEICNKIVEACENWGIFQVVDHGVDSKLISEMTRFAKGFFDLPPE 354
Query: 63 HRFKEVISS--KGLDVVQTEVKD---MDWES--TFHVRHLPESNISEIPDLSDEYRKVMK 115
+ + +S KG +V + ++ DW T+ + + + S PD + +++V +
Sbjct: 355 EKLRFDMSGGKKGGFIVSSHLQGEAVKDWRELVTYFSYPIRQRDYSRWPDKPEGWKEVTE 534
Query: 116 EFSLRLEKLAEELLDLLCENLGLEKGYLKKAFYGSRGPTFGTKVANYPQCPNPELVKGLR 175
++S +L LA +LL++L E +GLEK L KA + YP+CP P+L GL+
Sbjct: 535 QYSEKLMNLACKLLEVLSEAMGLEKDALTKACVDMDQKVV---INYYPKCPQPDLTLGLK 705
Query: 176 AHTDAGGIILLFQDDKVSGLQLLKDG--QWVDVPPMRHSIVVNLGDQLEVITNGKYKSVE 233
HTD G I LL QD +V GLQ KD W+ V P+ + VVNLGD ++NG++K+ +
Sbjct: 706 RHTDPGTITLLLQD-QVGGLQATKDNGKTWITVQPVEGAFVVNLGDHGHYLSNGRFKNAD 882
Query: 234 HRVIAQTNGTRMSIASFYNPGSDAVIYP 261
H+ + +N +R+SIA+F NP DA +YP
Sbjct: 883 HQAVVNSNYSRLSIATFQNPAPDATVYP 966
>TC80530 weakly similar to PIR|S44261|S44261 SRG1 protein - Arabidopsis
thaliana, partial (52%)
Length = 1219
Score = 163 bits (413), Expect = 7e-41
Identities = 96/299 (32%), Positives = 165/299 (55%), Gaps = 10/299 (3%)
Frame = +2
Query: 5 PIISLEKL---NGVERKDTMEKIKDACENWGFFELVNHGIPHDLMDTLERLTKEHYRKCM 61
PII +KL +G+E +EK+ +AC+ WGFF+L+NHG+ L+++++ ++ + M
Sbjct: 137 PIIDFDKLLCEDGIE----LEKLDNACKEWGFFQLINHGVNPSLVESVKIGVQQFFHLPM 304
Query: 62 EHRFK-----EVISSKGLDVVQTEVKDMDWESTFHVRHLP-ESNISE-IPDLSDEYRKVM 114
E + K E + G V E + + W F+V+ P + IP + +R
Sbjct: 305 EEKKKFWQTEEELQGFGQVYVALEEQKLRWGDMFYVKTFPLHIRLPHLIPCMPQPFRDDF 484
Query: 115 KEFSLRLEKLAEELLDLLCENLGLEKGYLKKAFYGSRGPTFGTKVANYPQCPNPELVKGL 174
+ +SL L+KL ++++ + + L +++ F+ + ++ YP CP P+ V GL
Sbjct: 485 ENYSLELKKLCFKIIERMTKALKIQQPNELLDFFEEGDQSI--RMNYYPPCPQPDQVIGL 658
Query: 175 RAHTDAGGIILLFQDDKVSGLQLLKDGQWVDVPPMRHSIVVNLGDQLEVITNGKYKSVEH 234
H+DA + +L Q +++ GLQ+ KDG WV + P+ ++ VVN+GD LE++TNG Y+S+EH
Sbjct: 659 NPHSDASALTILLQVNEMQGLQIKKDGMWVPINPLPNAFVVNIGDLLEIMTNGIYRSIEH 838
Query: 235 RVIAQTNGTRMSIASFYNPGSDAVIYPAPELLEKQTEEKHNVYPKFVFEEYMKIYAALK 293
R A + R+S+A F+N + PAP L+ T E ++ EEY+ Y A K
Sbjct: 839 RATANSEKERISVAGFHNIQMGRDLGPAPSLV---TPETPAMFKTITLEEYVNGYLASK 1006
>TC91229 similar to GP|9294689|dbj|BAB03055.1 gene_id:MHC9.10~similar to
oxidases {Arabidopsis thaliana}, partial (86%)
Length = 1198
Score = 160 bits (404), Expect = 8e-40
Identities = 98/307 (31%), Positives = 161/307 (51%), Gaps = 12/307 (3%)
Frame = +2
Query: 3 NFPIISLEKLNGVERKDT---MEKIKDACENWGFFELVNHGIPHDLMDTLERLTKEHYRK 59
+ P+I K + +++ + K+ ACE WGFF+++NH I +LM+ +E +++E +
Sbjct: 161 DMPVIDFSKFSKGNKEEVFNELSKLSTACEEWGFFQVINHEIDLNLMENIEDMSREFFML 340
Query: 60 CMEHRFKE-----VISSKGLDVVQTEVKDMDWESTFHVRHLPE--SNISEIPDLSDEYRK 112
++ + K + G V +E + +DW + F + P N + P +
Sbjct: 341 PLDEKQKYPMAPGTVQGYGQAFVFSEDQKLDWCNMFALGIEPHYIRNSNLWPKKPARLSE 520
Query: 113 VMKEFSLRLEKLAEELLDLLCENLGLEKGYLKKAFYGSRGPTFGTKVANYPQCPNPELVK 172
++ +S ++ KL + LL + L LE+ +K F + ++ YP C P+LV
Sbjct: 521 TIELYSRKIRKLCQNLLKYIALGLSLEEDVFEKMFGEA---VQAIRMNYYPTCSRPDLVL 691
Query: 173 GLRAHTDAGGIILLFQDDKVS--GLQLLKDGQWVDVPPMRHSIVVNLGDQLEVITNGKYK 230
GL H+D G + + Q K S GLQ+LKD WV V P+ +++V+N+GD +EV+TNGKYK
Sbjct: 692 GLSPHSD-GSALTVLQQAKGSPVGLQILKDNTWVPVEPISNALVINIGDTIEVLTNGKYK 868
Query: 231 SVEHRVIAQTNGTRMSIASFYNPGSDAVIYPAPELLEKQTEEKHNVYPKFVFEEYMKIYA 290
SVEHR +A R+SI +FY P D + P EL+++ K Y ++ EY K Y
Sbjct: 869 SVEHRAVAHEEKDRLSIVTFYAPSYDLELGPMLELVDENHPCK---YKRYNHGEYSKHYV 1039
Query: 291 ALKFHAK 297
K K
Sbjct: 1040TNKLQGK 1060
>TC89553 weakly similar to SP|P24397|HY6H_HYONI Hyoscyamine 6-dioxygenase
(EC 1.14.11.11) (Hyoscyamine 6-beta- hydroxylase).
[Henbane], partial (49%)
Length = 1119
Score = 158 bits (400), Expect = 2e-39
Identities = 101/310 (32%), Positives = 156/310 (49%), Gaps = 9/310 (2%)
Frame = +1
Query: 7 ISLEKLNGVERKDTMEKIKDACENWGFFELVNHGIPHDLMDTLERLTKEHYRKCMEHRFK 66
I L L G + T+ +I A E +GFF+++NHG+ DLMD + +E + + +
Sbjct: 130 IPLIDLGGHDHAHTILQILKASEEYGFFQVINHGVSKDLMDEAFNIFQEFHAMPPKEKIS 309
Query: 67 EVISS-KGLDV---VQTEVKDMD----WESTFHVRHLPESNISEI-PDLSDEYRKVMKEF 117
E G++ +E +D W+ T P E P +YR+V+ ++
Sbjct: 310 ECSRDPNGINCKIYASSENYKIDAVQYWKDTLTHPCPPSGEFMEFWPQKPPKYREVVGKY 489
Query: 118 SLRLEKLAEELLDLLCENLGLEKGYLKKAFYGSRGPTFGTKVANYPQCPNPELVKGLRAH 177
+ L KL E+L++LCE LGL+ Y F G ++P CP P L GL H
Sbjct: 490 TQELNKLGHEILEMLCEGLGLKLEY----FIGELSENPVILGHHFPPCPEPSLTLGLAKH 657
Query: 178 TDAGGIILLFQDDKVSGLQLLKDGQWVDVPPMRHSIVVNLGDQLEVITNGKYKSVEHRVI 237
D I +L QD +V GLQ+LKD +W+ V P+ +++VVN+G L++ITNG+ EHRV+
Sbjct: 658 RDPTIITILLQDQEVHGLQILKDDEWIPVEPIPNALVVNIGLILQIITNGRLVGAEHRVV 837
Query: 238 AQTNGTRMSIASFYNPGSDAVIYPAPELLEKQTEEKHNVYPKFVFEEYMKIYAALKFHAK 297
+ R S+A F P +I PA EL++ +Y F E+ K KF+ K
Sbjct: 838 TNSKSVRTSVAYFIYPSFSRMIEPAKELVDGNNPP---IYKSMSFGEFRK-----KFYDK 993
Query: 298 EPRFEALKES 307
P+ E + +S
Sbjct: 994 GPKIEQVLQS 1023
>TC78109 similar to PIR|S44261|S44261 SRG1 protein - Arabidopsis thaliana,
partial (48%)
Length = 1314
Score = 109 bits (273), Expect(2) = 3e-39
Identities = 64/212 (30%), Positives = 113/212 (53%), Gaps = 12/212 (5%)
Frame = +3
Query: 5 PIISLEKLNGVE--RKDTMEKIKDACENWGFFELVNHGIPHDLMDTLERLTKEHYRKCME 62
P+I L KL + ++K+ AC+ WGFF+L+NHG+ LM+ +++ +E + ME
Sbjct: 195 PVIDLSKLLSSHDLNEPELKKLHYACKEWGFFQLINHGVSESLMENVKKGAEEFFNLPME 374
Query: 63 HRFK-----EVISSKGLDVVQTEVKDMDWESTFHVRHLPESN-----ISEIPDLSDEYRK 112
+ K + G V +E + +DW + LP S IP +R
Sbjct: 375 EKKKFGQTEGDVEGYGQAFVVSEEQKLDWADMLVLFTLPPHKRKPHLFSNIPL---PFRV 545
Query: 113 VMKEFSLRLEKLAEELLDLLCENLGLEKGYLKKAFYGSRGPTFGTKVANYPQCPNPELVK 172
++ + ++ LA +++DL+ +L ++ +++ F+G T T++ YP CP PE V
Sbjct: 546 DLENYCEKMRTLAIQIMDLMAHSLAVDPMEIRE-FFGQA--TQSTRMNYYPPCPQPEFVI 716
Query: 173 GLRAHTDAGGIILLFQDDKVSGLQLLKDGQWV 204
GL +H+D GG+ +L Q +++ GLQ+ KDG W+
Sbjct: 717 GLNSHSDGGGLTILLQGNEMDGLQIKKDGLWI 812
Score = 70.1 bits (170), Expect(2) = 3e-39
Identities = 34/92 (36%), Positives = 54/92 (57%)
Frame = +1
Query: 206 VPPMRHSIVVNLGDQLEVITNGKYKSVEHRVIAQTNGTRMSIASFYNPGSDAVIYPAPEL 265
V P+ ++ ++NLGD LE++TNG ++S+EHR + R SIASFY+P + ++ PAP +
Sbjct: 817 VKPLPNAFIINLGDMLEMMTNGIFRSIEHRATVNSEKERFSIASFYSPAFNTILSPAPSI 996
Query: 266 LEKQTEEKHNVYPKFVFEEYMKIYAALKFHAK 297
+ T V+ + EY K Y A + K
Sbjct: 997 VTPNTPA---VFKRISAGEYFKGYLAQELCGK 1083
>TC86474 weakly similar to GP|6016680|gb|AAF01507.1| putative
leucoanthocyanidin dioxygenase {Arabidopsis thaliana},
partial (20%)
Length = 1395
Score = 153 bits (387), Expect = 7e-38
Identities = 92/281 (32%), Positives = 150/281 (52%), Gaps = 11/281 (3%)
Frame = +1
Query: 18 KDTMEKIKDACENWGFFELVNHGIPHDLMDTLERLTKEHYRKCMEHRFK-----EVISSK 72
K+ + K+ AC+ WGFF++VNHG+ DLM L+ + E + +E + K + I
Sbjct: 274 KNELLKLDIACKEWGFFQIVNHGMEIDLMQRLKDVVAEFFDLSIEEKDKYAMPPDDIQGY 453
Query: 73 GLDVVQTEVKDMDWESTFHVRHLPES--NISEIPDLSDEYRKVMKEFSLRLEKLAEELLD 130
G V +E + +DW + P P+ ++ + ++ +S ++++ EEL++
Sbjct: 454 GHTSVVSEKQILDWCDQLILLVYPTRFRKPQFWPETPEKLKDTIEAYSSEIKRVGEELIN 633
Query: 131 LLCENLGLEK----GYLKKAFYGSRGPTFGTKVANYPQCPNPELVKGLRAHTDAGGIILL 186
L GLE+ G K+ G R V YP C PE V GL H+DA + ++
Sbjct: 634 SLSLIFGLEEHVLLGLHKEVLQGLR-------VNYYPPCNTPEQVIGLTPHSDASTVTIV 792
Query: 187 FQDDKVSGLQLLKDGQWVDVPPMRHSIVVNLGDQLEVITNGKYKSVEHRVIAQTNGTRMS 246
QDD V+GL++ G WV + P+ +++VVNLGD +EV++NGKYKSVEHR + N R S
Sbjct: 793 MQDDDVTGLEVRYKGNWVPINPIPNALVVNLGDVIEVLSNGKYKSVEHRAMTNKNKRRTS 972
Query: 247 IASFYNPGSDAVIYPAPELLEKQTEEKHNVYPKFVFEEYMK 287
SF P DA + P +++ Q + +Y + + EY++
Sbjct: 973 FVSFLFPRDDAELGPFDHMIDDQNPK---MYKEITYGEYLR 1086
>BF650974 similar to SP|P31239|ACCO 1-aminocyclopropane-1-carboxylate oxidase
(EC 1.-.-.-) (ACC oxidase) (Ethylene-forming enzyme)
(EFE), partial (40%)
Length = 582
Score = 149 bits (376), Expect = 1e-36
Identities = 67/115 (58%), Positives = 86/115 (74%)
Frame = +1
Query: 1 MMNFPIISLEKLNGVERKDTMEKIKDACENWGFFELVNHGIPHDLMDTLERLTKEHYRKC 60
M FP++ ++ LN E+ TME IKDACENWGFFELVNHGI + MD +E+LTKE+Y+K
Sbjct: 193 MEKFPVVDMKNLNTEEKASTMEIIKDACENWGFFELVNHGISIEFMDKVEKLTKENYKKR 372
Query: 61 MEHRFKEVISSKGLDVVQTEVKDMDWESTFHVRHLPESNISEIPDLSDEYRKVMK 115
ME R KE+++SKGL Q+E+ D+DWESTF +RHLP NIS IPDL +YR +
Sbjct: 373 MEQRXKEMVASKGLXFAQSEINDLDWESTFFLRHLPNFNISXIPDLDHDYRSYQR 537
Score = 37.7 bits (86), Expect = 0.006
Identities = 16/19 (84%), Positives = 18/19 (94%)
Frame = +2
Query: 169 ELVKGLRAHTDAGGIILLF 187
+L+KGLR HTDAGGIILLF
Sbjct: 524 DLIKGLRXHTDAGGIILLF 580
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.318 0.137 0.403
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,109,368
Number of Sequences: 36976
Number of extensions: 117747
Number of successful extensions: 821
Number of sequences better than 10.0: 173
Number of HSP's better than 10.0 without gapping: 701
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 705
length of query: 317
length of database: 9,014,727
effective HSP length: 96
effective length of query: 221
effective length of database: 5,465,031
effective search space: 1207771851
effective search space used: 1207771851
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 58 (26.9 bits)
Medicago: description of AC137995.4


