

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137995.11 + phase: 0
(1094 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
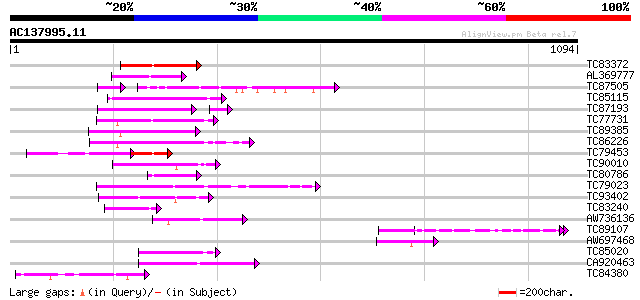
Score E
Sequences producing significant alignments: (bits) Value
TC83372 weakly similar to GP|20339360|gb|AAM19353.1 TIR-similar-... 117 2e-26
AL369777 weakly similar to GP|20339360|gb TIR-similar-domain-con... 92 8e-19
TC87505 weakly similar to PIR|T05981|T05981 hypothetical protein... 74 7e-18
TC85115 similar to GP|19774147|gb|AAL99050.1 NBS-LRR-Non-Toll re... 86 6e-17
TC87193 similar to GP|21616922|gb|AAM66423.1 NBS-LRR protein {Or... 72 3e-16
TC77731 weakly similar to GP|18033111|gb|AAL56987.1 functional c... 81 3e-15
TC89385 similar to GP|19774173|gb|AAL99063.1 NBS-LRR-Toll resist... 79 1e-14
TC86226 weakly similar to GP|18033111|gb|AAL56987.1 functional c... 77 3e-14
TC79453 weakly similar to GP|20339360|gb|AAM19353.1 TIR-similar-... 77 5e-14
TC90010 similar to GP|5817349|gb|AAD52718.1| putative NBS-LRR ty... 76 8e-14
TC80786 similar to GP|20339360|gb|AAM19353.1 TIR-similar-domain-... 74 4e-13
TC79023 weakly similar to GP|18033111|gb|AAL56987.1 functional c... 73 5e-13
TC93402 weakly similar to GP|16588610|gb|AAL26857.1 NBS-type put... 70 6e-12
TC83240 similar to GP|20339360|gb|AAM19353.1 TIR-similar-domain-... 66 6e-11
AW736136 weakly similar to GP|6625522|emb| putative serine-threo... 65 1e-10
TC89107 weakly similar to GP|14994693|gb|AAK76992.1 kruppel like... 64 2e-10
AW697468 64 3e-10
TC85020 similar to GP|19774173|gb|AAL99063.1 NBS-LRR-Toll resist... 64 4e-10
CA920463 weakly similar to GP|2852684|gb|A resistance protein ca... 64 4e-10
TC84380 62 1e-09
>TC83372 weakly similar to GP|20339360|gb|AAM19353.1
TIR-similar-domain-containing protein TSDC {Pisum
sativum}, partial (21%)
Length = 1151
Score = 117 bits (294), Expect = 2e-26
Identities = 62/158 (39%), Positives = 97/158 (61%), Gaps = 1/158 (0%)
Frame = +1
Query: 214 ERIQEKIAGSLEFEFQEKDEMDRSKRLCMRLTQEDR-VLVILDDVWQMLDFDAIGIPSIE 272
+RIQ++IA L EF E R++++ + L DR +LVILDDV + LD + +GIP
Sbjct: 1 KRIQDEIANELNLEFDVNTEAGRTRKIYLTLANMDRQILVILDDVSENLDPEKVGIPCNS 180
Query: 273 HHKGCKILITSRSEAVCTLMDCQKKIQLSTLTNDETWDLFQKQALISEGTWISIKNMARE 332
+ CK+L+T+ + C + CQ++IQLS L+ +E W LF+K + I + +KN+A
Sbjct: 181 NR--CKVLLTTCRQQDCEFIHCQREIQLSPLSTEEAWTLFKKHSGIDNESSSDLKNVAYN 354
Query: 333 ISNECKGLPVATVAVASSLKGKAEVEWKVALDRLRSSK 370
++ EC+GLP + SSL+ K EWK +LD L+ S+
Sbjct: 355 VAIECEGLPRTIIDAGSSLRSKPIEEWKASLDHLKYSR 468
>AL369777 weakly similar to GP|20339360|gb TIR-similar-domain-containing
protein TSDC {Pisum sativum}, partial (31%)
Length = 497
Score = 92.4 bits (228), Expect = 8e-19
Identities = 51/149 (34%), Positives = 83/149 (55%), Gaps = 4/149 (2%)
Frame = +1
Query: 197 NLFDQVLFVPISSTVEVERIQEKIAGSLEFEFQEKDEMDRSKRLCMRLTQEDR-VLVILD 255
N+FD++LFV ++ + + ++IA L E R++++ + DR +LVI D
Sbjct: 46 NIFDEILFVNVTKNPNITAMHDEIADFLNIRLDRNSETGRARKILSTIEDMDRPILVIFD 225
Query: 256 DVWQMLDFDAIGIPSIEHHKGCKILITSRSEAVCTLMDCQKKIQLSTLTNDETWDLFQKQ 315
DV D +GIP + CK+L+T+R + C LM CQ++I L L+ +E LF+K
Sbjct: 226 DVRAKFDLRDVGIPC--NSNLCKVLLTARRQKYCDLMHCQREILLDPLSTEEASTLFEKH 399
Query: 316 ALISE---GTWISIKNMAREISNECKGLP 341
+ I E + + N+ARE++ EC GLP
Sbjct: 400 SGILEEDHSSSFDLFNVAREVAFECDGLP 486
>TC87505 weakly similar to PIR|T05981|T05981 hypothetical protein F17M5.60 -
Arabidopsis thaliana, partial (45%)
Length = 2140
Score = 73.9 bits (180), Expect(2) = 7e-18
Identities = 109/456 (23%), Positives = 191/456 (40%), Gaps = 67/456 (14%)
Frame = +3
Query: 247 EDRVLVILDDVWQMLDFDAIGIPSIEHHKGCKILITSRSEAVCTLMDCQKKIQLSTLTND 306
+ ++LVILDDVW + + + CK ++ SR + + + K++L L D
Sbjct: 387 QPQILVILDDVWSPSVLEQL----VFRMPNCKFIVVSRF--IFPIFNATYKVEL--LDKD 542
Query: 307 ETWDLFQKQALISEGT-WISIKNMAREISNECKGLPVATVAVASSLKGKAEVEWKVALDR 365
+ LF A + + + +N+ +++ EC LP+A + +SL+ + E+ W R
Sbjct: 543 DALSLFCHHAFGQKSIPFAANQNLVKQVVAECGNLPLALKVIGASLRDQNEMFWLSVKTR 722
Query: 366 LRSSKPVNIEKGLQNPYKCLQLSYDNLDTEEAKSLFL-LCSVFPEDCEIPVEFLTRSAIG 424
L S+ ++I++ + N E+ K FL LCS FPED +IP+E L +
Sbjct: 723 L--SQGLSIDESYERNLIDRMAISTNYLPEKIKECFLDLCS-FPEDKKIPLEVLINMWVE 893
Query: 425 LGIVGEVHSYE-----GARNEVTVAKNK----LISSCLLLDVNEGKCVKMHDLVRNVAHW 475
+ + E +Y +N +T+ + + SSC + V + HD++R++A
Sbjct: 894 IHDIHETEAYAIVVELSNKNLLTLVEEARAGGMYSSCFEISVTQ------HDILRDLALN 1055
Query: 476 IA-------ENEIKCASEKDIMTLEHTSLRYLWCEKFPNSL-----------DCSNLDF- 516
++ + +D L LRY + F + D NL+F
Sbjct: 1056LSNRGNINQRRRLVMPKREDNGQLPKEWLRYA-DQPFEAQIVSIHTGEMRKSDWCNLEFP 1232
Query: 517 -LQIHTYTQVSDEIF-----KGMRMLRVLFLYNKGRERRPLLTTSL-KSLTNLRCILFSK 569
++ S E F M LR L + N L S+ K+LTNLR + F K
Sbjct: 1233KAEVLIINFTSSEYFLPPFINRMPKLRALMVINHSTSYACLHNISVFKNLTNLRSLWFEK 1412
Query: 570 WDLVDIS--FVGDMKKL-------------------------ESITLCDCSFV-ELPDVV 601
+ +S + ++KL +TL C V ELP +
Sbjct: 1413VSIPHLSGIVMESLRKLFIVLCKINNSLEGKDSNIADIFPNISELTLDHCEDVTELPSSI 1592
Query: 602 TQLTNLRLLDLSEC-GMERNPFEV-IARHTELEELF 635
++ +L+ L L+ C + R P E+ R+ E+ L+
Sbjct: 1593CRIQSLQNLSLTNCHSLTRLPIELGSLRYLEILRLY 1700
Score = 35.8 bits (81), Expect(2) = 7e-18
Identities = 19/57 (33%), Positives = 34/57 (59%), Gaps = 3/57 (5%)
Frame = +2
Query: 170 DVTMIGLYGMGGCGKTMLAMEVGKR---CGNLFDQVLFVPISSTVEVERIQEKIAGS 223
D ++G+ G+GG GKT LA E+ + G +++LF+ +S + VE+ + K G+
Sbjct: 140 DFCLVGISGIGGSGKTTLAREICRDEQVRGYFKERILFLTVSQSPNVEQPEGKDLGT 310
Score = 33.1 bits (74), Expect = 0.60
Identities = 23/74 (31%), Positives = 35/74 (47%), Gaps = 1/74 (1%)
Frame = +3
Query: 548 RRPLLTTSLKSLTNLRCILFSKWDLVDISFVGDMKKLESITLCDCSFV-ELPDVVTQLTN 606
R P+ SL+ L LR + S G M +L+ I + C ++ PD + +L N
Sbjct: 1647 RLPIELGSLRYLEILRLYACPNLRTLPPSICG-MTRLKYIDIAQCVYLASFPDAIGKLVN 1823
Query: 607 LRLLDLSECGMERN 620
L +D+ EC M N
Sbjct: 1824 LEKIDMRECPMITN 1865
>TC85115 similar to GP|19774147|gb|AAL99050.1 NBS-LRR-Non-Toll resistance
gene analog protein {Medicago ruthenica}, partial (90%)
Length = 746
Score = 86.3 bits (212), Expect = 6e-17
Identities = 65/236 (27%), Positives = 118/236 (49%), Gaps = 7/236 (2%)
Frame = +3
Query: 190 EVGKRCGNLFDQVLFVPISSTVEVERIQEKIAGSLEFEFQ--EKDEMDRSKRLCMRLTQE 247
E+GKR + FD VL+ +S + ++I I+ + + E+ D+ +E
Sbjct: 27 ELGKR-EHSFDLVLWAVVSKDCDTKKIMTDISRRIGIDDSVWEESSQDQKVGKIYEGLKE 203
Query: 248 DRVLVILDDVWQMLDFDAIGIPSI-EHHKGCKILITSRSEAVCTLMDCQKKIQLSTLTND 306
+ L++LDD+W+ L+ +G P + E + K++ T+R + VC M QKK+++ L+++
Sbjct: 204 KKFLLMLDDLWEKLELQELGFPVLKESNNKSKVVFTTRIKDVCAKMKAQKKLEVKCLSDE 383
Query: 307 ETWDLFQ-KQALISEGTWISIKNMAREISNECKGLPVATVAVASSLKGKAEVE-WKVALD 364
E ++LF+ K + I +A E++ +C+GLP+A + V S++ G E W+VA +
Sbjct: 384 EAFELFRSKLGDETLNCHTEIPKLAHEMAKKCRGLPLALITVGSAMAGVESYEAWEVAKN 563
Query: 365 RLR--SSKPVNIEKGLQNPYKCLQLSYDNLDTEEAKSLFLLCSVFPEDCEIPVEFL 418
L S +I K P L+ N K+ + +D EI V+ L
Sbjct: 564 NLSNCSWTAGDIVKVFSYP---SSLAMTNFQIRHIKAASCIVHYTQKDYEIDVDEL 722
>TC87193 similar to GP|21616922|gb|AAM66423.1 NBS-LRR protein {Oryza sativa
(japonica cultivar-group)}, partial (26%)
Length = 3119
Score = 72.0 bits (175), Expect(2) = 3e-16
Identities = 55/198 (27%), Positives = 106/198 (52%), Gaps = 8/198 (4%)
Frame = +3
Query: 170 DVTMIGLYGMGGCGKTMLAMEVGK--RCGNLFDQVLFVPISSTVEVERIQEKIAGSLEFE 227
+V+++ + G+GG GKT LA V K NLF++ ++V +S + + I + + SL E
Sbjct: 642 NVSLVAIVGIGGLGKTALAQLVYKDGEVKNLFEKHMWVCVSDNFDFKTILKNMVASLTKE 821
Query: 228 -FQEKDEMDRSKRLCMRLTQEDRVLVILDDVW--QMLDFDAIGIPSIEHHKGCKILITSR 284
K + L + LT + R L++LDDVW +D + + +G K+++T+
Sbjct: 822 DVVNKTLQELQSMLQVNLTGK-RYLLVLDDVWNESFEKWDQLRPYLMCGAQGSKVVMTTC 998
Query: 285 SEAVCTLMDCQKKIQLSTLTNDETWDLFQKQAL--ISEGTWISIKNMAREISNECKGLPV 342
S+ V M + L LT +++W LF+ ++ G ++++ ++I+ +CKG+P+
Sbjct: 999 SKIVADRMGVSDQHVLRGLTPEKSWVLFKNIVFGDVTVGVNQPLESIGKKIAEKCKGVPL 1178
Query: 343 ATVAVASSLKGKA-EVEW 359
A ++ L+ ++ E EW
Sbjct: 1179AIRSLGGILRSESKESEW 1232
Score = 33.5 bits (75), Expect = 0.46
Identities = 37/137 (27%), Positives = 66/137 (48%), Gaps = 5/137 (3%)
Frame = +2
Query: 482 KCASEKDIMTLEHTSLRYLWCEKFPNSLDCSNLDFLQIHTYTQVSDE-----IFKGMRML 536
KC + +E+ +L L SLD S L L + +Y +F+ ++ L
Sbjct: 1634 KCLGRPVHVLVEYDALCLL------ESLDSSRLRTLIVMSYNDYKFRRPEISVFRNLKYL 1795
Query: 537 RVLFLYNKGRERRPLLTTSLKSLTNLRCILFSKWDLVDISFVGDMKKLESITLCDCSFVE 596
R L + + R+ + LK L +L + + + S +G++ L++I L D S V+
Sbjct: 1796 RFLKMESCSRQGVGFIE-KLKHLRHLDLRNYDSGESLSKS-IGNLVCLQTIKLND-SVVD 1966
Query: 597 LPDVVTQLTNLRLLDLS 613
P+VV++L NLR L ++
Sbjct: 1967 SPEVVSKLINLRHLTIN 2017
Score = 32.0 bits (71), Expect(2) = 3e-16
Identities = 16/45 (35%), Positives = 26/45 (57%)
Frame = +1
Query: 385 LQLSYDNLDTEEAKSLFLLCSVFPEDCEIPVEFLTRSAIGLGIVG 429
L+LSY NL ++ + F CS+FP+D E + L + + G +G
Sbjct: 1300 LKLSYQNLSPQQ-RQCFAYCSLFPQDWEFEKDELIQMWMAQGYLG 1431
>TC77731 weakly similar to GP|18033111|gb|AAL56987.1 functional candidate
resistance protein KR1 {Glycine max}, partial (23%)
Length = 1651
Score = 80.9 bits (198), Expect = 3e-15
Identities = 63/242 (26%), Positives = 124/242 (51%), Gaps = 7/242 (2%)
Frame = +1
Query: 168 DDDVTMIGLYGMGGCGKTMLAMEVGKRCGNLFDQVLFV----PISSTVEVERIQEKIAGS 223
+D+V M+GL+G GG GK+ LA V + F+ V F+ S+ ++ +Q+K+
Sbjct: 637 EDEVRMVGLFGTGGMGKSTLAKAVYNFVADQFEGVCFLHNVRENSTHKNLKHLQKKLLSK 816
Query: 224 L-EFEFQEKDEMDRSKRLCMRLTQEDRVLVILDDVWQMLDFDAI--GIPSIEHHKGCKIL 280
+ +F+ + +D + + RL+++ ++L+ILDDV ++ +A+ G+ H G +++
Sbjct: 817 IVKFDGKLEDVSEGIPIIKERLSRK-KILLILDDVDKLEQLEALAGGLDWFGH--GSRVI 987
Query: 281 ITSRSEAVCTLMDCQKKIQLSTLTNDETWDLFQKQALISEGTWISIKNMAREISNECKGL 340
IT+R + + + L E +L ++ A ++ + + + + GL
Sbjct: 988 ITTRDKHLLACHGITSTHAVEELNETEALELLRRMAFKNDKVPSTYEEILNRVVTYASGL 1167
Query: 341 PVATVAVASSLKGKAEVEWKVALDRLRSSKPVNIEKGLQNPYKCLQLSYDNLDTEEAKSL 400
P+A V + +L G+ +WK LD + +I+ + LQ+SYD L+ +E KS+
Sbjct: 1168PLAIVTIGDNLFGRKVEDWKRILDEYENIPNKDIQ-------RILQVSYDALEPKE-KSV 1323
Query: 401 FL 402
FL
Sbjct: 1324FL 1329
>TC89385 similar to GP|19774173|gb|AAL99063.1 NBS-LRR-Toll resistance gene
analog protein {Medicago ruthenica}, partial (98%)
Length = 1285
Score = 79.0 bits (193), Expect = 1e-14
Identities = 63/222 (28%), Positives = 105/222 (46%), Gaps = 7/222 (3%)
Frame = +3
Query: 153 DSRKPAYEELMCALKDDDVTMIGLYGMGGCGKTMLAMEVGKRCGNLFDQVLFVPISSTV- 211
+SR A + + D V +IG++GMGG GKT LAM + + + FD F+ S +
Sbjct: 606 NSRTEALKHQLLLNSVDGVRVIGIWGMGGKGKTTLAMNLYGQICHRFDASCFIDDVSKIF 785
Query: 212 -----EVERIQEKIAGSLEFEFQEKDEMDRSKRLCMRLTQEDRVLVILDDVWQMLDFDAI 266
++ ++ + +L E + + L ++ L+ILD+V Q+ + I
Sbjct: 786 RLHDGPIDAQKQILHQTLGIEHHQICNHYSATDLIRHRLSREKTLLILDNVDQVEQLERI 965
Query: 267 GIPSIEHHKGCKILITSRSEAVCTLMDCQKKIQLSTLTNDETWDLFQKQALISEGTWI-S 325
G+ G +I+I SR E + ++ L E+ LF+++A E + +
Sbjct: 966 GVHREWLGAGSRIVIISRDEHILKEYKVDVDYKVPLLDWTESHKLFRQKAFKLEKIIMKN 1145
Query: 326 IKNMAREISNECKGLPVATVAVASSLKGKAEVEWKVALDRLR 367
+N+A EI N GLP+ + S L G+ EWK AL RLR
Sbjct: 1146YQNLAYEILNYANGLPLTITVLGSFLSGRNVTEWKSALARLR 1271
>TC86226 weakly similar to GP|18033111|gb|AAL56987.1 functional candidate
resistance protein KR1 {Glycine max}, partial (24%)
Length = 1769
Score = 77.4 bits (189), Expect = 3e-14
Identities = 82/326 (25%), Positives = 146/326 (44%), Gaps = 7/326 (2%)
Frame = +3
Query: 154 SRKPAYEELMCALKDDDVTMIGLYGMGGCGKTMLAMEVGKRCGNLFDQVLFV----PISS 209
SR + L+ DD V M+G+YG+GG GK+ LA + + F+ + F+ S+
Sbjct: 639 SRVQQVKSLLDEGPDDGVHMVGIYGIGGSGKSTLARAIYNFVADQFEGLCFLEQVRENSA 818
Query: 210 TVEVERIQEK-IAGSLEFEFQEKDEMDRSKRLCMRLTQEDRVLVILDDVWQMLDFDAIGI 268
+ ++R QE ++ +L+ + + D + + RL ++ ++L+ILDDV M +A+
Sbjct: 819 SNSLKRFQEMLLSKTLQLKIKLADVSEGISIIKERLCRK-KILLILDDVDNMKQLNALAG 995
Query: 269 PSIEHHKGCKILITSRSEAVCTLMDCQKKIQLSTLTNDETWDLFQKQALISEGTWISIKN 328
G +++IT+R + + + +K + L E +L + A ++ S +
Sbjct: 996 GVDWFGPGSRVIITTRDKHLLACHEIEKTYAVKGLNVTEALELLRWMAFKNDKVPSSYEK 1175
Query: 329 MAREISNECKGLPVATVAVASSLKGKAEVEWKVALDRLRSSKPVNIEKGLQNPYKCLQLS 388
+ + GLPV V S+L GK E K LD I+ + L++S
Sbjct: 1176ILNRVVAYASGLPVVIEIVGSNLFGKNIEECKNTLDWYEKIPNKEIQ-------RILKVS 1334
Query: 389 YDNLDTEEAKSLFLLCSVFPEDC--EIPVEFLTRSAIGLGIVGEVHSYEGARNEVTVAKN 446
YD+L+ EE +S+FL + + C E E L H + V V
Sbjct: 1335YDSLE-EEEQSVFLDIACCFKGCKWEKVKEIL-----------HAHYGHCINHHVEV--- 1469
Query: 447 KLISSCLLLDVNEGKCVKMHDLVRNV 472
L+ CL+ V +H+L+ N+
Sbjct: 1470-LVEKCLIDHFEYDSHVSLHNLIENM 1544
>TC79453 weakly similar to GP|20339360|gb|AAM19353.1
TIR-similar-domain-containing protein TSDC {Pisum
sativum}, partial (38%)
Length = 975
Score = 76.6 bits (187), Expect = 5e-14
Identities = 57/212 (26%), Positives = 102/212 (47%), Gaps = 2/212 (0%)
Frame = +1
Query: 33 VQDLAKEESNLAAIRDSVQDRVTRAKKQTRKTAEVVEKWLKDANIAMDNVDQLLQMAKSE 92
++D+ +E+ L + DSVQ+++ +T+K ++V +WLK+ + V+ + + + E
Sbjct: 178 IKDVEREKKKLISNHDSVQEKIEATDHKTQKVNDIVLEWLKEVEKLVQEVENVTIIPEPE 357
Query: 93 KNSCFGHCPNWIWRYSVGRKLSKKKRNLKLYIEEGRQYIEIERPASLSAGYFSAERCWEF 152
RY K+ K + L + E + I S+G F F
Sbjct: 358 S------------RYP--NKMLNKLKALNIKCEFEPFFNPIPSLEHFSSGNFVC-----F 480
Query: 153 DSRKPAYEELMCALKDDDVTMIGLYGMGGCGKTMLAMEVGKRCGNL--FDQVLFVPISST 210
+ K + L+ AL++ IGLYG G GKT L V ++ L F VLF+ +S
Sbjct: 481 EPIKETSDRLLEALENRKFYKIGLYGKRGSGKTKLVKAVAEKARYLRVFAAVLFITVSQN 660
Query: 211 VEVERIQEKIAGSLEFEFQEKDEMDRSKRLCM 242
V++IQ++IA L+ +F + E+ +S R+ +
Sbjct: 661 PNVKQIQDEIADFLDLKFDKNTEVGKS*RIIL 756
Score = 68.9 bits (167), Expect = 1e-11
Identities = 34/83 (40%), Positives = 55/83 (65%), Gaps = 1/83 (1%)
Frame = +2
Query: 233 EMDRSKRLCMRLTQEDR-VLVILDDVWQMLDFDAIGIPSIEHHKGCKILITSRSEAVCTL 291
+++R++ L + L DR +LVILDDVW+ LD + +GIP + CK+L+T+ + L
Sbjct: 728 KLERARELYLTLESTDRPILVILDDVWENLDLEELGIPCNSNR--CKVLLTTHCKQEFAL 901
Query: 292 MDCQKKIQLSTLTNDETWDLFQK 314
M+CQ++I L L+ +E W LF+K
Sbjct: 902 MNCQEEIPLCPLSIEEAWTLFKK 970
>TC90010 similar to GP|5817349|gb|AAD52718.1| putative NBS-LRR type disease
resistance protein {Pisum sativum}, partial (88%)
Length = 1107
Score = 75.9 bits (185), Expect = 8e-14
Identities = 55/216 (25%), Positives = 101/216 (46%), Gaps = 7/216 (3%)
Frame = +3
Query: 199 FDQVLFVPISSTVEVERIQEKIAGSLEFEFQEKDEMDRSKRLCMRLTQEDRVLVILDDVW 258
FD +V +S ++ R+ + + S+ + +D + ++++E R L + DD+W
Sbjct: 27 FDLKAWVCVSEDFDIMRVTKSLLESVTSTTWDSKNLDVLRVELKKISREKRFLFVFDDLW 206
Query: 259 Q--MLDFDAIGIPSIEHHKGCKILITSRSEAVCTLMDCQKKIQLSTLTNDETWDLFQKQA 316
D++ + P I G ++IT+R + V + +L L+N++ W L K A
Sbjct: 207 NDNYNDWNELASPFINGKPGSMVIITTRQQKVAEVAHTFPIHKLKLLSNEDCWSLLSKHA 386
Query: 317 LISE----GTWISIKNMAREISNECKGLPVATVAVASSLKGKAEV-EWKVALDRLRSSKP 371
L S+ + +++ R+I+ +C GLP+A + L+ K ++ EW L +S
Sbjct: 387 LGSDEFHHSSNTALEETGRKIARKCGGLPIAAKTIGGLLRSKVDITEWTSIL----NSNI 554
Query: 372 VNIEKGLQNPYKCLQLSYDNLDTEEAKSLFLLCSVF 407
N+ N L LSY L + K F CS+F
Sbjct: 555 WNLRN--DNILPALHLSYQYLPS-HLKRCFAYCSIF 653
>TC80786 similar to GP|20339360|gb|AAM19353.1 TIR-similar-domain-containing
protein TSDC {Pisum sativum}, partial (20%)
Length = 831
Score = 73.6 bits (179), Expect = 4e-13
Identities = 42/107 (39%), Positives = 63/107 (58%), Gaps = 3/107 (2%)
Frame = +1
Query: 266 IGIPSIEHHKGCKILITSRSEAVCTLMDCQKKIQLSTLTNDETWDLFQKQALISE---GT 322
+GIPS + CK+L+T+R + C L+ CQ+KI L L+ +E LF+K + I E +
Sbjct: 1 VGIPSNSNR--CKVLLTARRQKYCDLVYCQRKILLDPLSTEEASTLFEKYSGILEEDHSS 174
Query: 323 WISIKNMAREISNECKGLPVATVAVASSLKGKAEVEWKVALDRLRSS 369
+ N+AREI+ EC GLP + SS++ K EW+ +L LR S
Sbjct: 175 SFDLFNVAREIAFECDGLPGKIIKEGSSVRSKPMEEWEKSLHNLRHS 315
>TC79023 weakly similar to GP|18033111|gb|AAL56987.1 functional candidate
resistance protein KR1 {Glycine max}, partial (37%)
Length = 2072
Score = 73.2 bits (178), Expect = 5e-13
Identities = 104/443 (23%), Positives = 193/443 (43%), Gaps = 10/443 (2%)
Frame = +3
Query: 168 DDDVTMIGLYGMGGCGKTMLAMEVGKRCGNLFDQVLF---VPISSTVE-VERIQEK-IAG 222
DD+V M+GLYG GG GK+ LA + + F+ + F V ++ST + ++ +QEK +
Sbjct: 828 DDEVHMVGLYGTGGIGKSTLAKAIYNFIADQFEVLCFLENVRVNSTSDNLKHLQEKLLLK 1007
Query: 223 SLEFEFQEKDEMDRSKRLCMRLTQEDRVLVILDDVWQMLDFDAIGIPSIEHHKGCKILIT 282
++ + + + RL ++ ++L+ILDDV ++ +A+ G +++IT
Sbjct: 1008 TVRLDIKLGGVSQGIPIIKQRLCRK-KILLILDDVDKLDQLEALAGGLDWFGPGSRVIIT 1184
Query: 283 SRSEAVCTLMDCQKKIQLSTLTNDETWDLFQKQALISEGTWISIKNMAREISNECKGLPV 342
+R++ + + + + L E +L + A E S +++ GLP+
Sbjct: 1185 TRNKHLLKIHGIESTHAVEGLNATEALELLRWMA-FKENVPSSHEDILNRALTYASGLPL 1361
Query: 343 ATVAVASSLKGKAEVEWKVALDRLRSSKPVNIEKGLQNPYKCLQLSYDNLDTEEAKSLFL 402
A V + S+L G++ + LD I+ + L++SYD+L+ EE +S+FL
Sbjct: 1362 AIVIIGSNLVGRSVQDSMSTLDGYEEIPNKEIQ-------RILKVSYDSLEKEE-QSVFL 1517
Query: 403 LCSVFPEDCEIP-VEFLTRSAIGLGIVGEVHSYEGARNEVTVAKNKLISSCLLLDVNEGK 461
+ + C+ P V+ + + G IV + V V K + L D
Sbjct: 1518 DIACCFKGCKWPEVKEILHAHYGHCIV----------HHVAVLAEKSLMDHLKYD----S 1655
Query: 462 CVKMHDLVRNVAHWIAENEIKC-ASEKDIMTLEHTSLRYLWCEKFPNSLDCSNLDFLQIH 520
V +HDL+ ++ + E E+ + E + L + N+ F +
Sbjct: 1656 YVTLHDLIEDMGKEVVRQESPDEPGERSRLWFERDIVHVLKKNTGTRKIKMINMKFPSME 1835
Query: 521 TYTQVSDEIFKGMRMLRVLFLYNKGRERRPL--LTTSLKSLTNLRCILFSKWDLVDISFV 578
+ + F+ M L+ F+ G + L L +SL+ + CI S
Sbjct: 1836 SDIDWNGNAFEKMTNLKT-FITENGHHSKSLEYLPSSLRVMKG--CIPKSPSSSSSNKKF 2006
Query: 579 GDMKKLESITLCDCSFV-ELPDV 600
DMK L L +C ++ +PDV
Sbjct: 2007 EDMKVL---ILNNCEYLTHIPDV 2066
>TC93402 weakly similar to GP|16588610|gb|AAL26857.1 NBS-type putative
resistance protein {Glycine max}, partial (61%)
Length = 676
Score = 69.7 bits (169), Expect = 6e-12
Identities = 58/231 (25%), Positives = 113/231 (48%), Gaps = 9/231 (3%)
Frame = +2
Query: 171 VTMIGLYGMGGCGKTMLAMEV--GKRCGNLFDQVLFVPISSTVEVERIQEKIAGSLEFEF 228
V+ I + G+GG GKT LA V +R F+ +V +S +V + I
Sbjct: 5 VSTISIVGLGGMGKTTLAQLVYNDRRIQETFELKAWVYVSEYFDVIGLTNAILRKFGTAE 184
Query: 229 QEKDEMDRSKRLCMRLTQEDRVLVILDDVWQMLD--FDAIGIPSIEHHKGCKILITSRSE 286
+D +L +LT ++ L+++DDVW++ + ++ + +P G KI++T+R +
Sbjct: 185 NSEDLDLLQWQLQKKLTGKN-YLLVMDDVWKLNEESWEKLLLPFNYGSSGSKIILTTRDK 361
Query: 287 AVCTLMDCQKKIQLSTLTNDETWDLFQKQAL----ISEGTWISIKNMAREISNECKGLPV 342
V ++ + + L L N + W LF++ A +SE + ++++ + I ++C GLP+
Sbjct: 362 KVALIVKSTELVDLEQLKNKDCWSLFKRLAFHGSNVSE--YPKLESIGKNIVDKCGGLPL 535
Query: 343 ATVAVASSLKGK-AEVEWKVALDRLRSSKPVNIEKGLQNPYKCLQLSYDNL 392
+ + L+ K + EW +++ + + N L+LSY NL
Sbjct: 536 TGKTMGNLLRKKFTQSEW----EKILEADMWRLTDDDSNINSALRLSYHNL 676
>TC83240 similar to GP|20339360|gb|AAM19353.1 TIR-similar-domain-containing
protein TSDC {Pisum sativum}, partial (23%)
Length = 337
Score = 66.2 bits (160), Expect = 6e-11
Identities = 40/112 (35%), Positives = 65/112 (57%), Gaps = 3/112 (2%)
Frame = +3
Query: 184 KTMLAMEVGKRCGNL--FDQVLFVPISSTVEVERIQEKIAGSLEFEFQEKDEMDRSKRLC 241
KT LA +G++ +L F +VLF ++ + + +QE+IA L+ F +K E R++R+
Sbjct: 3 KTALAKAMGEKVKHLKIFHEVLFATVTQNLNIRTMQEEIADLLDMTFDKKSETVRARRIF 182
Query: 242 MRLTQEDR-VLVILDDVWQMLDFDAIGIPSIEHHKGCKILITSRSEAVCTLM 292
R+ R +LVI DDV D + +GIP + CKIL+T+ ++ C LM
Sbjct: 183 SRIESMSRPILVIFDDVRVKFDPEDVGIPC--NSNRCKILLTALAQQDCELM 332
>AW736136 weakly similar to GP|6625522|emb| putative serine-threonine-protein
kinase {Schizosaccharomyces pombe}, partial (3%)
Length = 642
Score = 65.5 bits (158), Expect = 1e-10
Identities = 59/205 (28%), Positives = 95/205 (45%), Gaps = 22/205 (10%)
Frame = -3
Query: 276 GCKILITSRSEAVCTLMDCQKK--IQLSTLTN-------------DETWDLFQKQALISE 320
G K+++T+R + VC MD Q IQ+ L DE W+LF + E
Sbjct: 622 GIKVILTTRLKHVCHQMDSQTNDIIQMFPLCCLKESEDEVDEDKVDEGWELFMLKLGHDE 443
Query: 321 GTWI---SIKNMAREISNECKGLPVATVAVASSLKGKAEV-EWKVALDRLRSSKPVNIEK 376
I+ + R I KGLP+ +A ++ G ++ +WK AL RL+ + +
Sbjct: 442 TPRTLPHEIEEIVRCIVERFKGLPLGINLMARTMDGNDDIHQWKHALSRLQK---FEMRQ 272
Query: 377 GLQNPYKCLQLSYDNLDTEEAKSLFLLCSVFPEDCE---IPVEFLTRSAIGLGIVGEVHS 433
++ +K L+ SYDNL ++ ++ FL C++F D E I + L + G + E S
Sbjct: 271 VMEEVFKVLKCSYDNLMEKDLQNCFLYCALFSIDDEGWKINKDELIMKLVDNGQINENMS 92
Query: 434 YEGARNEVTVAKNKLISSCLLLDVN 458
EG +E +KL S L+ N
Sbjct: 91 LEGIFDEGNTIPSKLESH*LIKSTN 17
>TC89107 weakly similar to GP|14994693|gb|AAK76992.1 kruppel like factor 12
{Danio rerio}, partial (6%)
Length = 1361
Score = 64.3 bits (155), Expect = 2e-10
Identities = 75/305 (24%), Positives = 129/305 (41%), Gaps = 9/305 (2%)
Frame = -1
Query: 782 FENLEDLYISHCPKLTRLFTLAVAQNLAQLEKLQVLSCPELQHILIDDDRDEISAYDYRL 841
F+N+ L I+ C L+ +F L+VA+ L L++L + C + D+ D + +
Sbjct: 1331 FQNIFQLKITDCKGLSHVFPLSVAKELLHLQELYIEKCGIEIIVAQDETADTVP-----V 1167
Query: 842 LLFPKLKKFHVRECGVLEYIIPITLAQGLVQLECLEIVCNENLKYVFGQSTHNDGQNQNE 901
L FP+L R+ L GL L+CL
Sbjct: 1166 LNFPELTSLSFRDLTQLRRFY-----LGLHTLDCLF------------------------ 1074
Query: 902 LKIIELSALEELTLVNLPNINSICPEDCYLMWPSLLQFN--LQNCGEFFMVSINT---CM 956
LK +++ ++L L L ++N C ++ + LL + N E + S + C
Sbjct: 1073 LKDVDVLHCDKLELFTLRSLN--CQDNVLVDTLPLLSIEKVVSNTRELILNSKDVTMLCN 900
Query: 957 ALHNNPRIN---EASHQTLQNITEVRVNNCELEGIFQLVGLTNDG-EKDPLTSCLEMLYL 1012
HNN I + + T +++ + L + L + + E + +E L++
Sbjct: 899 GQHNNETIYTIFSGCASSGHSETIMKLRSLVLVNLHNLKFICEEKFEVQTVLQNIENLFV 720
Query: 1013 ENLPQLRYLCKSSVESTNLLFQNLQQMEISGCRRLKCIFSSCMAGGLPQLKALKIEKCNQ 1072
P+L + SSV LF+NLQQ+E+ C L+ I S A L +L+ L IE C +
Sbjct: 719 YRCPRLNNIVPSSV-----LFENLQQLEVGNCAGLENIVKSSTAISLQKLRKLIIEGCEK 555
Query: 1073 LDQIV 1077
+ +IV
Sbjct: 554 IGEIV 540
Score = 59.3 bits (142), Expect = 8e-09
Identities = 91/365 (24%), Positives = 163/365 (43%), Gaps = 7/365 (1%)
Frame = -1
Query: 712 GAKNIIP-DVFQSMNHLKELLIRDSKGIECLV--DTCLIEVGTLFFCKLHWLRIEHMKHL 768
G ++ P V + + HL+EL I + GIE +V D V L F +L L + L
Sbjct: 1292 GLSHVFPLSVAKELLHLQELYI-EKCGIEIIVAQDETADTVPVLNFPELTSLSFRDLTQL 1116
Query: 769 GALYNGQMPLSGHFENLEDLYISHCPKLTRLFTLAV--AQNLAQLEKLQVLSCPELQHIL 826
Y G L F L+D+ + HC KL LFTL Q+ ++ L +LS ++
Sbjct: 1115 RRFYLGLHTLDCLF--LKDVDVLHCDKL-ELFTLRSLNCQDNVLVDTLPLLSIEKV---- 957
Query: 827 IDDDRDEI-SAYDYRLLLFPKLKKFHVRECGVLEYIIPITLAQGLVQLECLEIVCNENLK 885
+ + R+ I ++ D +L + + + ++ +++L L +V NLK
Sbjct: 956 VSNTRELILNSKDVTMLCNGQHNNETIYT--IFSGCASSGHSETIMKLRSLVLVNLHNLK 783
Query: 886 YVFGQSTHNDGQNQNELKIIELSALEELTLVNLPNINSICPEDCYLMWPSLLQFNLQNCG 945
++ + + E++ + L +E L + P +N+I P S+L NLQ
Sbjct: 782 FIC--------EEKFEVQTV-LQNIENLFVYRCPRLNNIVPS-------SVLFENLQQ-- 657
Query: 946 EFFMVSINTCMALHNNPRINEASHQTLQNITEVRVNNCELEGIFQLVGLTNDGEKDPLTS 1005
+ + C L N + + A +LQ + ++ + CE G ++V ++ + L+
Sbjct: 656 ----LEVGNCAGLENIVKSSTAI--SLQKLRKLIIEGCEKIG--EIVASDDENDDSELSF 501
Query: 1006 C-LEMLYLENLPQLRYLCKSSVESTNLLFQNLQQMEISGCRRLKCIFSSCMAGGLPQLKA 1064
LE L L NLP+LR CK L F LQ++ + C ++ + P+L+A
Sbjct: 500 MKLEYLRLSNLPRLRSFCKG---RHGLKFPLLQKLFVVDCPMMETFSHGVL--NAPKLRA 336
Query: 1065 LKIEK 1069
L +++
Sbjct: 335 LNVKE 321
>AW697468
Length = 653
Score = 63.9 bits (154), Expect = 3e-10
Identities = 46/146 (31%), Positives = 65/146 (44%), Gaps = 28/146 (19%)
Frame = +1
Query: 709 IEGGAKNIIPD---VFQSMNHLKELLIRDSKGIECLVDTCLIEVGTLFFCKLHWLRIEHM 765
+ GG KNIIPD + MN L L + + IEC+ D +L LR+ HM
Sbjct: 1 LHGGCKNIIPDMVGIVGGMNDLTSLHLTSCQEIECIFDATYDFKEDDLIPRLGELRLRHM 180
Query: 766 KHLGALYNG-------------------------QMPLSGHFENLEDLYISHCPKLTRLF 800
+L LY G PL NL+ L +S+C LF
Sbjct: 181 NNLTELYRGPSLQVLRYFEKLELVDIQYCWQVHIMFPLECKLRNLKILSLSNCRTDKVLF 360
Query: 801 TLAVAQNLAQLEKLQVLSCPELQHIL 826
+ +VAQ++ QLE+L + C EL+HI+
Sbjct: 361 SESVAQSMLQLEQLNISGCYELKHII 438
>TC85020 similar to GP|19774173|gb|AAL99063.1 NBS-LRR-Toll resistance gene
analog protein {Medicago ruthenica}, partial (55%)
Length = 500
Score = 63.5 bits (153), Expect = 4e-10
Identities = 51/160 (31%), Positives = 82/160 (50%), Gaps = 1/160 (0%)
Frame = +1
Query: 249 RVLVILDDVWQMLDFDAIGIPSIEHHKGCKILITSRSEAVCTLMDCQKKIQLSTLTNDET 308
+ L+I D+V Q+ + I + G +I+I SR E + ++ + + ++
Sbjct: 1 KALLIFDNVDQVEQLEKIAVHREWLGAGSRIVIISRDEHILKEYGVDVVYKVPLMNSTDS 180
Query: 309 WDLFQKQALISEGTWIS-IKNMAREISNECKGLPVATVAVASSLKGKAEVEWKVALDRLR 367
++LF ++A E +S +N+A EI + KGLP+A + S L G + EWK AL RLR
Sbjct: 181 YELFCRKAFKVEKIIMSDYQNLANEILDYAKGLPLAIKVLGSFLFGHSVAEWKSALARLR 360
Query: 368 SSKPVNIEKGLQNPYKCLQLSYDNLDTEEAKSLFLLCSVF 407
S P N + L LS+D LD E+ K +FL + F
Sbjct: 361 ES-PHN------DVMDVLHLSFDGLD-EKEKEIFLDIACF 456
>CA920463 weakly similar to GP|2852684|gb|A resistance protein candidate
{Lactuca sativa}, partial (6%)
Length = 810
Score = 63.5 bits (153), Expect = 4e-10
Identities = 63/242 (26%), Positives = 115/242 (47%), Gaps = 8/242 (3%)
Frame = -1
Query: 249 RVLVILDDVWQMLDFDAIGIPSIEH--HKGCKILITSRSEAVCTLMDCQKKIQLSTLTND 306
+ L+ LDDVW I + ++ +G K+L+T+RS ++ +M L L+ +
Sbjct: 753 KFLLRLDDVWSEDRVKWIEVKNLLQVGDEGSKVLVTTRSHSIAKMMCTNTSYTLQGLSRE 574
Query: 307 ETWDLFQKQALIS--EGTWISIKNMAREISNECKGLPVATVAVASSLKGKAEV-EWKVAL 363
++ +F K A E + + + +EI +C GLP+A + SSL K ++ EWK
Sbjct: 573 DSLSVFVKWAFKEGEEKKYPKLIEIGKEIVQKCGGLPLALRTLGSSLFLKDDIEEWKF-- 400
Query: 364 DRLRSSKPVNIEKGLQNPYKCLQLSYDNLDTEEAKSLFLLCSVFPEDCEIPVEFLTRSAI 423
+R ++ N+ + + L+LS+D L + K F S+F +D +T
Sbjct: 399 --VRDNEIWNLPQKEDDILPALKLSFDQLPS-YLKRCFACFSLFVKDFHFSNYSVTVLWE 229
Query: 424 GLGIVGEVHSYEGARNEVTVAKNKLISSCLLLD--VNEGKCV-KMHDLVRNVAHWIAENE 480
L + + + + ++L S L D V+ CV K+HDLV ++A ++A +E
Sbjct: 228 ALDFLPSPNKGKTLEDVGNQFLHELQSRSFLQDFYVSGNVCVFKLHDLVHDLALYVARDE 49
Query: 481 IK 482
+
Sbjct: 48 FQ 43
>TC84380
Length = 761
Score = 62.0 bits (149), Expect = 1e-09
Identities = 72/277 (25%), Positives = 115/277 (40%), Gaps = 18/277 (6%)
Frame = +1
Query: 11 ISRDLVCGVIGQLSYPCCFNNFVQDLAKEESNLAAIRDSVQDRVTRAKKQTRKTAEVVEK 70
I R ++CG G C F+ ++ E + + + + K +T K +VV +
Sbjct: 7 ILRSILCGFTGL----CLFSILDYFVSMENQSSVSSLTLLVLKKVEEKNKTEKINDVVME 174
Query: 71 WLKDAN----------IAMDNVDQLLQMAKSEKNSCFGHCPNWIWRYSVGRKLSKKKRNL 120
WL D + I M+ ++ L SEK RY + ++ +K + L
Sbjct: 175 WLNDVDKVMEEEEKMEIEMEILEILCTSIDSEK------------RYRLYNEMLRKIKTL 318
Query: 121 KLYIEEGRQYIEIERPASLSAG--YFSAERCWEFDSRKPAYEELMCALKDDDVTMIGLYG 178
E E +S G YFS F+S K A ++L+ AL+D + +IGLYG
Sbjct: 319 NTNCE-------FEPFSSPIPGLEYFSFGNFVCFESTKVASDQLLEALQDGNCYIIGLYG 477
Query: 179 MGGCGKTMLAMEVGKRCG--NLFDQVLFVPISSTVEVERIQEKIAGSLEF----EFQEKD 232
G GKT L VG++ +FD V IS V + + E
Sbjct: 478 KKGSGKTKLVKAVGEKAKYLKIFDA---VSISQCVSKSKC*NNSR*NCPITEP*NLTEIP 648
Query: 233 EMDRSKRLCMRLTQEDRVLVILDDVWQMLDFDAIGIP 269
+++ ++ DR+LVIL+DV L+ + I IP
Sbjct: 649 KLEEQEQYLRHYKVGDRILVILNDVCSKLELEDISIP 759
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.322 0.137 0.411
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 37,187,033
Number of Sequences: 36976
Number of extensions: 557024
Number of successful extensions: 3036
Number of sequences better than 10.0: 182
Number of HSP's better than 10.0 without gapping: 2886
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2990
length of query: 1094
length of database: 9,014,727
effective HSP length: 106
effective length of query: 988
effective length of database: 5,095,271
effective search space: 5034127748
effective search space used: 5034127748
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 64 (29.3 bits)
Medicago: description of AC137995.11


