

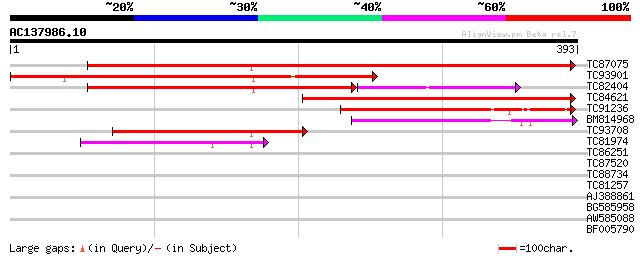
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137986.10 - phase: 0
(393 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC87075 similar to GP|16209674|gb|AAL14395.1 AT5g15050/F2G14_170... 406 e-114
TC93901 similar to GP|6091754|gb|AAF03464.1| hypothetical protei... 317 7e-87
TC82404 similar to GP|9294673|dbj|BAB03022.1 gb|AAF26009.1~gene_... 187 3e-68
TC84621 similar to GP|10176991|dbj|BAB10223. glycosylation enzym... 214 5e-56
TC91236 similar to PIR|D85042|D85042 probable glycosylation enzy... 187 6e-48
BM814968 similar to PIR|B96735|B96 unknown protein F23N20.6 [imp... 136 1e-32
TC93708 weakly similar to PIR|B96735|B96735 unknown protein F23N... 121 5e-28
TC81974 weakly similar to GP|20197252|gb|AAM14996.1 putative RIN... 79 3e-15
TC86251 similar to GP|22136176|gb|AAM91166.1 unknown protein {Ar... 31 0.94
TC87520 similar to GP|9757998|dbj|BAB08420.1 cell division prote... 31 0.94
TC88734 similar to GP|13346180|gb|AAK19612.1 GHDEL61 {Gossypium ... 30 1.6
TC81257 similar to GP|9279781|dbj|BAB01433.1 glucosyltransferase... 30 1.6
AJ388861 weakly similar to GP|1754989|gb| proline-rich protein P... 30 1.6
BG585958 weakly similar to GP|22830897|dbj ORF-B {Oryza sativa (... 29 2.7
AW585088 similar to GP|14278913|dbj NADH dehydrogenase subunit 6... 28 6.1
BF005790 similar to PIR|T47763|T477 hypothetical protein F24I3.1... 28 8.0
>TC87075 similar to GP|16209674|gb|AAL14395.1 AT5g15050/F2G14_170
{Arabidopsis thaliana}, partial (67%)
Length = 1863
Score = 406 bits (1044), Expect = e-114
Identities = 192/345 (55%), Positives = 251/345 (72%), Gaps = 7/345 (2%)
Frame = +2
Query: 55 AYLISASKGDTLKLKRLLKVLYHPNNYYLIHMDYGAPDAEHKDVAEYVANDPVFSQVGNV 114
AYLIS S GD LKR LK LYHP N Y +H+D A E D+A +V N+P+F+++GNV
Sbjct: 347 AYLISGSVGDGESLKRTLKALYHPRNQYAVHLDLEASPKERLDLANFVRNEPLFAELGNV 526
Query: 115 WIVGKPNLVTYRGPTMLATTLHAMAMLLKTC-HWDWFINLSASDYPLVTQDGM------V 167
++ K NLVTYRGPTM+ TLHA A+L K WDWFINLSASDYPL+TQD + +
Sbjct: 527 RMIVKANLVTYRGPTMVTNTLHAAALLFKEAGDWDWFINLSASDYPLLTQDDLLHTLSSI 706
Query: 168 PRDINFIQHTSRLGWKFNKRGKPMIIDPGLYSLNKSDIWWIIKQRNLPTSFKLYTGSAWT 227
PR +NFI+HTS +GWK ++R KP+IIDP LYS+NKSD++W+ ++R++PT++KL+TGSAW
Sbjct: 707 PRHLNFIEHTSDIGWKEDQRAKPVIIDPALYSVNKSDVFWVTEKRSVPTAYKLFTGSAWM 886
Query: 228 IVSRSFSEYCIMGWENLPRTLLLYYTNFVSSPEGYFQTVICNSQEYKNTTANHDLHYITW 287
++SR F EY + GW+NLPR +L+YY NF+SSPEGYF TVICN++E++NTT NHDLH+I+W
Sbjct: 887 MLSRQFVEYMLWGWDNLPRIVLMYYANFLSSPEGYFHTVICNAEEFRNTTVNHDLHFISW 1066
Query: 288 DNPPKQHPRSLGLKDYRKMVLSSRPFARKFKRNNIVLDKIDRDLLKRYKGGFSFGGWCSQ 347
DNPPKQHP L + Y MV S+ PF RKF RN +LDKID +LL R G+ G W S
Sbjct: 1067DNPPKQHPHFLTAEHYWSMVESNAPFGRKFGRNEPLLDKIDTELLGRNADGYVPGMWFSH 1246
Query: 348 GGRNKACSGLRAENYGLLKPGPGSRRLKNLLNKILMDKFFRQMQC 392
N +N L+PGPG++RLKNL+N +L + FR+ QC
Sbjct: 1247ANPNITKPYSFVKNITELRPGPGAKRLKNLINGLLSAEDFRKNQC 1381
>TC93901 similar to GP|6091754|gb|AAF03464.1| hypothetical protein
{Arabidopsis thaliana}, partial (50%)
Length = 826
Score = 317 bits (811), Expect = 7e-87
Identities = 160/267 (59%), Positives = 196/267 (72%), Gaps = 12/267 (4%)
Frame = +1
Query: 1 MGIKIFMFTFMVTSILFFFLFIPTRLTLQISTLKPS--AMDYFNVLRTNITYPITFAYLI 58
M IKIF+ ++TS LFF L+IPT+LT+ S P FN+ TN YPI+FAYLI
Sbjct: 31 MDIKIFIIASILTSTLFFVLYIPTKLTIPTSNYSPPNPMNKTFNISNTNKKYPISFAYLI 210
Query: 59 SASKGDTLKLKRLLKVLYHPNNYYLIHMDYGAPDAEHKDVAEYVANDPVFSQVGNVWIVG 118
S SKGD +K+KRLLK LYHP NYYLIHMD+GAP +H+DV +YV+N+PVF QVGNVWI+
Sbjct: 211 SGSKGDAIKIKRLLKALYHPGNYYLIHMDHGAPKKDHRDVIDYVSNEPVFGQVGNVWIIK 390
Query: 119 KPNLVTYRGPTMLATTLHAMAMLLKTCHWDWFINLSASDYPLVTQDGMV---------PR 169
K N VTY+GPTML+TTLHAMA+LL++C WDWFINLSASDYPLVTQDGM+
Sbjct: 391 KSNFVTYKGPTMLSTTLHAMAILLRSCKWDWFINLSASDYPLVTQDGMI*SMPFLEFQSI 570
Query: 170 DINFIQHTSRLGWKFNKRGKPMIIDPGLYSLNKSDIWWI-IKQRNLPTSFKLYTGSAWTI 228
I+FI +G + + + I Y ++ I+W KQR+LPT+FKLYTGSAWT+
Sbjct: 571 LISFIITVIWVGNSIREGSQYL*IQH--YIAKRNQIYWQGTKQRSLPTAFKLYTGSAWTM 744
Query: 229 VSRSFSEYCIMGWENLPRTLLLYYTNF 255
+SRSFSEYCI+GWENLPR LLLYYTNF
Sbjct: 745 LSRSFSEYCIIGWENLPRILLLYYTNF 825
>TC82404 similar to GP|9294673|dbj|BAB03022.1
gb|AAF26009.1~gene_id:F14O13.23~similar to unknown
protein {Arabidopsis thaliana}, partial (65%)
Length = 925
Score = 187 bits (474), Expect(2) = 3e-68
Identities = 85/193 (44%), Positives = 140/193 (72%), Gaps = 7/193 (3%)
Frame = +2
Query: 55 AYLISASKGDTLKLKRLLKVLYHPNNYYLIHMDYGAPDAEHKDVAEYVANDPVFSQVGNV 114
AYLIS SKGD+ ++ RLL YHP N YL+H+D AP ++ + +A V ++ VF NV
Sbjct: 2 AYLISGSKGDSGRILRLLYATYHPLNQYLLHLDPSAPQSDREKLALVVQSNTVFEAAKNV 181
Query: 115 WIVGKPNLVTYRGPTMLATTLHAMAMLLK-TCHWDWFINLSASDYPLVTQDGMV------ 167
++GKP+ V +G + ++ TLHA A+L++ + WDWF++LSA YPLVTQD ++
Sbjct: 182 HVMGKPDFVYVKGSSPVSFTLHAAAILIRLSLRWDWFVSLSADSYPLVTQDDLLHIMSFL 361
Query: 168 PRDINFIQHTSRLGWKFNKRGKPMIIDPGLYSLNKSDIWWIIKQRNLPTSFKLYTGSAWT 227
P+D+NF+ H+S +GWK +K+ KP+I+DPGLY +++++ ++R LP++++++TGS+++
Sbjct: 362 PKDMNFVNHSSYIGWKESKKLKPIIVDPGLYLSEGTEMFYATQKRELPSAYRMFTGSSFS 541
Query: 228 IVSRSFSEYCIMG 240
I+SR+F E+CI+G
Sbjct: 542 ILSRNFMEFCILG 580
Score = 90.1 bits (222), Expect(2) = 3e-68
Identities = 46/113 (40%), Positives = 66/113 (57%)
Frame = +3
Query: 242 ENLPRTLLLYYTNFVSSPEGYFQTVICNSQEYKNTTANHDLHYITWDNPPKQHPRSLGLK 301
+NLPR LL+Y++N SS YF TV+CNS+++ T N DL Y +D+ + + R L
Sbjct: 585 DNLPRILLMYFSNTPSSLSNYFPTVLCNSRQFNKTVINQDLLYAVYDS-HRNNLRPLNST 761
Query: 302 DYRKMVLSSRPFARKFKRNNIVLDKIDRDLLKRYKGGFSFGGWCSQGGRNKAC 354
D+ M+ S FA+KF+ ++ VLD ID+ +L R G GGWC N C
Sbjct: 762 DFDNMIHSGAVFAKKFQPDDPVLDLIDQKILNRTPGSVVPGGWCLGEPGNSTC 920
>TC84621 similar to GP|10176991|dbj|BAB10223. glycosylation enzyme-like
protein {Arabidopsis thaliana}, partial (42%)
Length = 709
Score = 214 bits (545), Expect = 5e-56
Identities = 98/189 (51%), Positives = 134/189 (70%)
Frame = +2
Query: 204 DIWWIIKQRNLPTSFKLYTGSAWTIVSRSFSEYCIMGWENLPRTLLLYYTNFVSSPEGYF 263
D++W+ ++R+ PT+F L+TGSAW ++SR F +Y I GW+NLPRT+L+YY+NF+SSPEGYF
Sbjct: 5 DVFWVTQRRSRPTAF*LFTGSAWMVLSRPFVDYVIWGWDNLPRTVLMYYSNFISSPEGYF 184
Query: 264 QTVICNSQEYKNTTANHDLHYITWDNPPKQHPRSLGLKDYRKMVLSSRPFARKFKRNNIV 323
TVICN+QE++NTT N DLH+I WDNPPKQHP L + D + M S+ PFARKF R + V
Sbjct: 185 HTVICNAQEFRNTTVNSDLHFIAWDNPPKQHPHYLTVADMKVMTDSNAPFARKFHREDPV 364
Query: 324 LDKIDRDLLKRYKGGFSFGGWCSQGGRNKACSGLRAENYGLLKPGPGSRRLKNLLNKILM 383
LD+ID +LL R G GGWC + N N +L+P GS+RL+ L+ K++
Sbjct: 365 LDRIDTELLSRNPGMPVPGGWCIRSRENGTDPCSVVGNTTVLRPENGSKRLETLITKLMS 544
Query: 384 DKFFRQMQC 392
++ FR QC
Sbjct: 545 NENFRPRQC 571
>TC91236 similar to PIR|D85042|D85042 probable glycosylation enzyme
[imported] - Arabidopsis thaliana, partial (35%)
Length = 788
Score = 187 bits (475), Expect = 6e-48
Identities = 95/165 (57%), Positives = 120/165 (72%), Gaps = 2/165 (1%)
Frame = +3
Query: 230 SRSFSEYCIMGWENLPRTLLLYYTNFVSSPEGYFQTVICNSQEYKNTTANHDLHYITWDN 289
+RSF EYCI GW+N PRT+L+YYTNFVSSPEGYF TVICN+Q++++T +HDLHYI WD+
Sbjct: 3 TRSFVEYCIWGWDNFPRTMLMYYTNFVSSPEGYFHTVICNTQKFRHTAISHDLHYIAWDS 182
Query: 290 PPKQHPRSLGLKDYRKMVLSSRPFARKFKRNNIVLDKIDRDLLKRYKGGFSFGGWC--SQ 347
PPKQHP SL +K++ KM S+ PFARKF R++ VLDKID++LL R FS G WC S
Sbjct: 183 PPKQHPMSLTMKNFDKMAKSNAPFARKFARDDPVLDKIDKELLGR-THRFSPGAWCIGSS 359
Query: 348 GGRNKACSGLRAENYGLLKPGPGSRRLKNLLNKILMDKFFRQMQC 392
G CS LR N + +PGPG+ +L LL +L D+ FR QC
Sbjct: 360 DGGADPCS-LRG-NDTVFRPGPGADKLHELLQVLLSDE-FRSKQC 485
>BM814968 similar to PIR|B96735|B96 unknown protein F23N20.6 [imported] -
Arabidopsis thaliana, partial (34%)
Length = 742
Score = 136 bits (343), Expect = 1e-32
Identities = 75/170 (44%), Positives = 99/170 (58%), Gaps = 14/170 (8%)
Frame = +2
Query: 238 IMGWENLPRTLLLYYTNFVSSPEGYFQTVICNSQEYKNTTANHDLHYITWDNPPKQHPRS 297
I GW+NLPRTLL+Y+TN S EGYF +VICN+ EYKNTT N DL Y+ WDNPPK P
Sbjct: 17 IFGWDNLPRTLLMYFTNVKLSQEGYFHSVICNAPEYKNTTVNGDLRYMIWDNPPKMEPLF 196
Query: 298 LGLKDYRKMVLSSRPFARKFKRNNIVLDKIDRDLLKRYKGGFSFGGWCSQGGRNKA---- 353
L Y M S FAR+F+ NN VLD ID+ +L+R GGRN+A
Sbjct: 197 LNTSVYDMMAESGAAFARQFEANNPVLDMIDKKILQR-------------GGRNRAAPGA 337
Query: 354 -CSGLRA---------ENYGLLKPGPGSRRLKNLLNKILMDKFFRQMQCR 393
CSG R+ + +LKPGP +++L+ ++ +L D + QC+
Sbjct: 338 WCSGRRSWWVDPCSQWGDVNILKPGPQAKKLEASVSSLLDDWTAQTNQCQ 487
>TC93708 weakly similar to PIR|B96735|B96735 unknown protein F23N20.6
[imported] - Arabidopsis thaliana, partial (49%)
Length = 991
Score = 121 bits (303), Expect = 5e-28
Identities = 62/142 (43%), Positives = 89/142 (62%), Gaps = 7/142 (4%)
Frame = +1
Query: 72 LKVLYHPNNYYLIHMDYGAPDAEHKDVAEYVANDPVFSQVGNVWIVGKPNLVTYRGPTML 131
L +YHP N YL+H+ A + E + +A+ V++ P GNV +VGK + +TY G + +
Sbjct: 550 LLAVYHPRNRYLLHLGMDARNEERQGLADAVSSVPAIRAFGNVDVVGKADWITYLGSSNV 729
Query: 132 ATTLHAMAMLLK-TCHWDWFINLSASDYPLVTQDGM------VPRDINFIQHTSRLGWKF 184
A TL A A++LK W+WFI LSA DYPL+TQD + V RD+NFI HT LGWK
Sbjct: 730 AITLRAAAIMLKLDSGWNWFITLSARDYPLITQDDLSHVFSSVNRDLNFIDHTGDLGWKE 909
Query: 185 NKRGKPMIIDPGLYSLNKSDIW 206
+ R +P+++DPG +S I+
Sbjct: 910 SDRFQPIVVDPGNIFSRESQIF 975
>TC81974 weakly similar to GP|20197252|gb|AAM14996.1 putative RING zinc
finger protein {Arabidopsis thaliana}, partial (21%)
Length = 654
Score = 79.0 bits (193), Expect = 3e-15
Identities = 47/138 (34%), Positives = 75/138 (54%), Gaps = 8/138 (5%)
Frame = +3
Query: 50 YPITFAYLISASKGDTLKLKRLLKVLYHPNNYYLIHMDYGAPDAEHKDVAEYVANDPVFS 109
YP FAY I +KG++ K+ RLLK +YHP N +L+ +D + D+E D+A Y+ + F
Sbjct: 180 YPPVFAYWIFGTKGESKKMMRLLKAIYHPRNQHLLQLDDCSSDSERMDLALYIKSHMGF* 359
Query: 110 QVGNVWIVGKPNLVTYRGPTMLATTLHAMA--MLLKTCHWDWFINLSASDYPLVTQDGM- 166
++ K + L+ + L T +WF LSASDYPL+TQD +
Sbjct: 360 RIWKCECCWKELCYQ*EWDLLHFLLLYMLLPYFLK*TQIGNWFFTLSASDYPLMTQDDIL 539
Query: 167 -----VPRDINFIQHTSR 179
+P+++NFI +T++
Sbjct: 540 HAFTSLPKNLNFIHYTNK 593
>TC86251 similar to GP|22136176|gb|AAM91166.1 unknown protein {Arabidopsis
thaliana}, partial (37%)
Length = 1636
Score = 30.8 bits (68), Expect = 0.94
Identities = 21/90 (23%), Positives = 36/90 (39%), Gaps = 4/90 (4%)
Frame = -3
Query: 6 FMFTFMVTSILFFFLFIPTRLTLQIST----LKPSAMDYFNVLRTNITYPITFAYLISAS 61
F FTF + FFF F + L L L P + + + + T+ T + S+S
Sbjct: 743 FSFTFSFSFTFFFFFFFSSSLPLSTPNVYFLLFPLSSPFPLISAVHFTFFTTALFFFSSS 564
Query: 62 KGDTLKLKRLLKVLYHPNNYYLIHMDYGAP 91
+ L L + P+ +H+ + P
Sbjct: 563 TNSAIHLYFLSNPFFSPSFISAVHVYFRPP 474
>TC87520 similar to GP|9757998|dbj|BAB08420.1 cell division protein FtsH
protease-like {Arabidopsis thaliana}, partial (53%)
Length = 1667
Score = 30.8 bits (68), Expect = 0.94
Identities = 12/25 (48%), Positives = 16/25 (64%)
Frame = -2
Query: 263 FQTVICNSQEYKNTTANHDLHYITW 287
F T+IC S+E+ N N LH+ TW
Sbjct: 1150 FHTIICISKEFNNLCINT*LHFRTW 1076
>TC88734 similar to GP|13346180|gb|AAK19612.1 GHDEL61 {Gossypium hirsutum},
partial (40%)
Length = 1701
Score = 30.0 bits (66), Expect = 1.6
Identities = 20/76 (26%), Positives = 35/76 (45%), Gaps = 3/76 (3%)
Frame = -1
Query: 208 IIKQRNLPTSFKLYTGSAWTIVSRSFSEYCIMGWENLPRTLLLYYTNFVSSPEGYFQTVI 267
+I + + L TG + V + FS+ + E+ + YY N + E VI
Sbjct: 1221 LILDESFENASPLLTGHCFLKVLKEFSDNANLHLEDPLNSFFGYYCNLLQP*EQKKYFVI 1042
Query: 268 C---NSQEYKNTTANH 280
C NS+ Y+++ +NH
Sbjct: 1041 CSHLNSRSYEHSCSNH 994
>TC81257 similar to GP|9279781|dbj|BAB01433.1 glucosyltransferase-like
protein {Arabidopsis thaliana}, partial (24%)
Length = 783
Score = 30.0 bits (66), Expect = 1.6
Identities = 21/81 (25%), Positives = 36/81 (43%), Gaps = 1/81 (1%)
Frame = +2
Query: 3 IKIFMFTFMVTSILFFFLFIP-TRLTLQISTLKPSAMDYFNVLRTNITYPITFAYLISAS 61
+ + FTF IL F +FIP + L L + P M + N+L + + P YL+ +
Sbjct: 29 LPFYSFTFFCI-ILPFPMFIPESELPLWVICYVPIVMSFLNILPSPKSVPFLVPYLLFEN 205
Query: 62 KGDTLKLKRLLKVLYHPNNYY 82
K ++ L+ + Y
Sbjct: 206 TMSVTKFNAMVSGLFQLGSAY 268
>AJ388861 weakly similar to GP|1754989|gb| proline-rich protein PRP2
precursor {Lupinus luteus}, partial (24%)
Length = 491
Score = 30.0 bits (66), Expect = 1.6
Identities = 11/13 (84%), Positives = 11/13 (84%)
Frame = +2
Query: 337 GGFSFGGWCSQGG 349
GGFS GGWCS GG
Sbjct: 239 GGFSCGGWCSGGG 277
>BG585958 weakly similar to GP|22830897|dbj ORF-B {Oryza sativa (japonica
cultivar-group)}, partial (13%)
Length = 769
Score = 29.3 bits (64), Expect = 2.7
Identities = 14/50 (28%), Positives = 27/50 (54%), Gaps = 2/50 (4%)
Frame = +1
Query: 3 IKIFMFTFMV--TSILFFFLFIPTRLTLQISTLKPSAMDYFNVLRTNITY 50
+ IF++ + T+ILF + + ++ LK +DYFN+L I++
Sbjct: 37 LSIFLYKLIK**TNILFTIIIVTNYFIFTVAYLKLGPLDYFNILTPFISF 186
>AW585088 similar to GP|14278913|dbj NADH dehydrogenase subunit 6 {Rana
nigromaculata}, partial (13%)
Length = 501
Score = 28.1 bits (61), Expect = 6.1
Identities = 16/37 (43%), Positives = 19/37 (51%)
Frame = +1
Query: 275 NTTANHDLHYITWDNPPKQHPRSLGLKDYRKMVLSSR 311
NTT NH T P RS+GL +RK+V SR
Sbjct: 73 NTTPNHTEQTPTLVRPNTGSRRSIGLLTFRKIVRCSR 183
>BF005790 similar to PIR|T47763|T477 hypothetical protein F24I3.110 -
Arabidopsis thaliana, partial (25%)
Length = 425
Score = 27.7 bits (60), Expect = 8.0
Identities = 17/60 (28%), Positives = 29/60 (48%), Gaps = 4/60 (6%)
Frame = -1
Query: 238 IMGWENLPRTLLL----YYTNFVSSPEGYFQTVICNSQEYKNTTANHDLHYITWDNPPKQ 293
++GW NL + LLL + ++ Y V+ S++Y T D H +T+ N +Q
Sbjct: 422 LLGWPNLLKLLLL*DQGRHQQCINLHFSYQTLVLVASRKYALTLVRMDAHIVTYWNS*QQ 243
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.325 0.140 0.447
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 14,120,999
Number of Sequences: 36976
Number of extensions: 227435
Number of successful extensions: 1813
Number of sequences better than 10.0: 32
Number of HSP's better than 10.0 without gapping: 1739
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1799
length of query: 393
length of database: 9,014,727
effective HSP length: 98
effective length of query: 295
effective length of database: 5,391,079
effective search space: 1590368305
effective search space used: 1590368305
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 59 (27.3 bits)
Medicago: description of AC137986.10


