

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137838.4 + phase: 0
(496 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
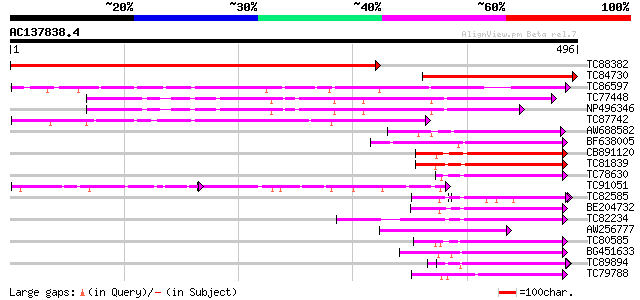
Score E
Sequences producing significant alignments: (bits) Value
TC88382 weakly similar to GP|6671958|gb|AAF23217.1| hypothetical... 670 0.0
TC84730 weakly similar to GP|6671958|gb|AAF23217.1| hypothetical... 270 9e-73
TC86597 weakly similar to GP|9837280|gb|AAG00510.1| leaf senesce... 182 2e-46
TC77448 SYMRK; MtSYMRK [Medicago truncatula]; nod region linked ... 154 6e-38
NP496346 NP496346|AJ418371.1|CAD10810.1 nod region linked recept... 145 3e-35
TC87742 weakly similar to GP|7269844|emb|CAB79703.1 serine/threo... 114 1e-25
AW688582 similar to GP|11072027|g F12A21.14 {Arabidopsis thalian... 110 1e-24
BF638005 weakly similar to GP|6560758|gb| F3M18.23 {Arabidopsis ... 105 4e-23
CB891120 similar to GP|17979065|gb putative receptor kinase homo... 99 5e-21
TC81839 similar to GP|21593619|gb|AAM65586.1 receptor protein ki... 97 1e-20
TC78630 similar to GP|10177052|dbj|BAB10464. receptor-like prote... 91 1e-18
TC91051 similar to PIR|H84787|H84787 probable receptor-like prot... 56 1e-18
TC82585 similar to GP|7715608|gb|AAF68126.1| F20B17.5 {Arabidops... 87 2e-17
BE204732 weakly similar to GP|8978275|db receptor protein kinase... 86 4e-17
TC82234 weakly similar to GP|7715608|gb|AAF68126.1| F20B17.5 {Ar... 85 7e-17
AW256777 weakly similar to PIR|T01269|T01 serine/threonine-speci... 82 6e-16
TC80585 weakly similar to GP|9759164|dbj|BAB09720.1 receptor kin... 82 6e-16
BG451633 weakly similar to GP|16930691|gb AT4g26540/M3E9_30 {Ara... 81 8e-16
TC89894 weakly similar to GP|14495542|gb|AAB36558.2 receptor-lik... 79 4e-15
TC79788 weakly similar to GP|21554189|gb|AAM63268.1 putative leu... 77 2e-14
>TC88382 weakly similar to GP|6671958|gb|AAF23217.1| hypothetical protein
{Arabidopsis thaliana}, partial (39%)
Length = 1207
Score = 670 bits (1729), Expect = 0.0
Identities = 324/324 (100%), Positives = 324/324 (100%)
Frame = +2
Query: 1 MSILFFLLLFISFHTPSFSQTPPKGFLINCGTLTTTQINNRTWLPDSNFITTGTPKNITT 60
MSILFFLLLFISFHTPSFSQTPPKGFLINCGTLTTTQINNRTWLPDSNFITTGTPKNITT
Sbjct: 236 MSILFFLLLFISFHTPSFSQTPPKGFLINCGTLTTTQINNRTWLPDSNFITTGTPKNITT 415
Query: 61 QVLLPTLKTLRSFPLQVKKHCYNIPVYRGAKYMIRTTYFYGGVNGVDHPTPPVFDQIIDG 120
QVLLPTLKTLRSFPLQVKKHCYNIPVYRGAKYMIRTTYFYGGVNGVDHPTPPVFDQIIDG
Sbjct: 416 QVLLPTLKTLRSFPLQVKKHCYNIPVYRGAKYMIRTTYFYGGVNGVDHPTPPVFDQIIDG 595
Query: 121 TLWSVVNTTVDYANGNSSFYEGVFLAVGKFMSFCIGSNSYTDSDPFVSALEFLILGDSLY 180
TLWSVVNTTVDYANGNSSFYEGVFLAVGKFMSFCIGSNSYTDSDPFVSALEFLILGDSLY
Sbjct: 596 TLWSVVNTTVDYANGNSSFYEGVFLAVGKFMSFCIGSNSYTDSDPFVSALEFLILGDSLY 775
Query: 181 NTTDFNNFAIGLVARNSFGYSGPSIRYPDDQFDRIWEPFGQSNSTKANTENVSVSGFWNL 240
NTTDFNNFAIGLVARNSFGYSGPSIRYPDDQFDRIWEPFGQSNSTKANTENVSVSGFWNL
Sbjct: 776 NTTDFNNFAIGLVARNSFGYSGPSIRYPDDQFDRIWEPFGQSNSTKANTENVSVSGFWNL 955
Query: 241 PPSKVFETHLGSEQLESLELRWPTASLPSSKYYIALYFADNTAGSRIFNISVNGVHYYRD 300
PPSKVFETHLGSEQLESLELRWPTASLPSSKYYIALYFADNTAGSRIFNISVNGVHYYRD
Sbjct: 956 PPSKVFETHLGSEQLESLELRWPTASLPSSKYYIALYFADNTAGSRIFNISVNGVHYYRD 1135
Query: 301 LNAIASGVVVFANQWPLSGPTTIT 324
LNAIASGVVVFANQWPLSGPTTIT
Sbjct: 1136LNAIASGVVVFANQWPLSGPTTIT 1207
>TC84730 weakly similar to GP|6671958|gb|AAF23217.1| hypothetical protein
{Arabidopsis thaliana}, partial (24%)
Length = 790
Score = 270 bits (690), Expect = 9e-73
Identities = 134/135 (99%), Positives = 135/135 (99%)
Frame = +1
Query: 362 RVKESLRNPPLDWSGDPCVPRQYSWTGITCSEGLRIRIVTLNLTSMDLSGSLSSFVANMT 421
+VKESLRNPPLDWSGDPCVPRQYSWTGITCSEGLRIRIVTLNLTSMDLSGSLSSFVANMT
Sbjct: 43 KVKESLRNPPLDWSGDPCVPRQYSWTGITCSEGLRIRIVTLNLTSMDLSGSLSSFVANMT 222
Query: 422 ALTNIWLGNNSLSGQIPNLSSLTMLETLHLEENQFSGEIPSSLGNISSLKEVFLQNNNLT 481
ALTNIWLGNNSLSGQIPNLSSLTMLETLHLEENQFSGEIPSSLGNISSLKEVFLQNNNLT
Sbjct: 223 ALTNIWLGNNSLSGQIPNLSSLTMLETLHLEENQFSGEIPSSLGNISSLKEVFLQNNNLT 402
Query: 482 GQIPANLLKPGLSIR 496
GQIPANLLKPGLSIR
Sbjct: 403 GQIPANLLKPGLSIR 447
>TC86597 weakly similar to GP|9837280|gb|AAG00510.1| leaf
senescence-associated receptor-like protein kinase
{Phaseolus vulgaris}, partial (46%)
Length = 2233
Score = 182 bits (463), Expect = 2e-46
Identities = 153/515 (29%), Positives = 245/515 (46%), Gaps = 26/515 (5%)
Frame = +3
Query: 2 SILFFLLLFISFHTPSFSQTPPKGFL-INCG-------TLTTTQINNRTWLPDSNFITTG 53
++L FLL+F GF+ ++CG + + T IN ++ D+ FI TG
Sbjct: 93 TVLHFLLVFFVL----IKAQDQSGFISLDCGLPENLNYSSSKTGIN---YISDAKFIDTG 251
Query: 54 TPKNI--TTQVLLPTLKTLRSFPLQVKKHCYNIPVYRGAKYMIRTTYFYGGVNGVDHPTP 111
K I T + L L+ +RSFP V+ +CYNI V G KY IRT++ YG + ++ P
Sbjct: 252 VSKRIARTDKFSLQQLEYVRSFPSGVR-NCYNISVTSGTKYFIRTSFLYGNYDNLNKP-- 422
Query: 112 PVFDQIIDGTLWSVVNTTVDYANGNSSFYEGVFLAVGKFMSFCIGSNSYTDSDPFVSALE 171
P FD +W V + + + +F E ++ ++ C+ + PF+SA+E
Sbjct: 423 PQFDLHFGANVWDTVKLSSNASL--FTFREIIYTPSLDYIQPCLVNTG--KGTPFISAIE 590
Query: 172 FLILGDSLYNTTDFNNFAIGLVARNSFGYSGPSIRYPDDQFDRIWEPFGQSN----STKA 227
L ++ Y T + + + RY DD FDRIW P+G + ST
Sbjct: 591 LRPLNNTAYVTYSAKSVLTNFFRLDVGSITNLEYRYKDDVFDRIWFPYGFIDWARLSTSL 770
Query: 228 NTENVSVSGFWNLPPSKVFETHLGSEQLES-LELRWPTASLPSSKYYIALYF------AD 280
N +++ + + PP+ V T + + L+ W T ++ + +YY +F A+
Sbjct: 771 NNDDLVQNDYE--PPATVMSTAVTPLNASAPLQFEWGTDNV-NDQYYTYFHFNEVEKLAE 941
Query: 281 NTAGSRIFNISVNGVHYYRDLNAIASGVVVFANQWPLSGPTT--ITLTPSASSSLGPLIN 338
N +R FN+SVNG H + + + PL+G + I+LT + +S+L P++N
Sbjct: 942 NE--TRAFNLSVNGNHRHGPVIPGYRETYTIFSPTPLTGAKSYQISLTKTENSTLPPILN 1115
Query: 339 AGEVFNVLSLG-GRTSTRDVIALQRVKESLRNPPLDWSGDPCVPRQYSWTGITCS-EGLR 396
A E++ V T DV + +K + +W GDPC P Y W G+ CS +G
Sbjct: 1116AFEIYKVKDFSQSETHQDDVDTITNIKNAY-GLTRNWEGDPCAPVNYVWEGLNCSIDGNN 1292
Query: 397 I-RIVTLNLTSMDLSGSLSSFVANMTALTNIWLGNNSLSGQIPNLSSLTMLETLHLEENQ 455
I RI++LNL+S L+G +SS ++S LTML+ L L N
Sbjct: 1293IPRIISLNLSSSGLTGEISS-----------------------SISKLTMLQYLDLSNNS 1403
Query: 456 FSGEIPSSLGNISSLKEVFLQNNNLTGQIPANLLK 490
SG +P L + SLK + + N LTG +P+ LL+
Sbjct: 1404LSGPLPDFLMQLRSLKVLNVGKNKLTGLVPSGLLE 1508
>TC77448 SYMRK; MtSYMRK [Medicago truncatula]; nod region linked receptor
kinase [Medicago truncatula]
Length = 3568
Score = 154 bits (390), Expect = 6e-38
Identities = 126/429 (29%), Positives = 200/429 (46%), Gaps = 18/429 (4%)
Frame = +1
Query: 68 KTLRSFPLQVKKHCYNIPVYRGAKYMIRTTYFYGGVNGVDHPTPPVFDQIIDGTLWSVVN 127
K +R F + K CYN+P + Y+IR + + +N + + V + G L S
Sbjct: 613 KNVRKFEIYEGKRCYNLPTVKDQVYLIRGIFPFDSLNSSFYVSIGVTEL---GELRSSRL 783
Query: 128 TTVDYANGNSSFYEGVFLAVGKFMSFCIGSNSYTDSDPFVSALEFLILGDSLYNTTDFNN 187
++ EGVF A ++ FC+ D +PF+S +E L + + F
Sbjct: 784 EDLEI--------EGVFRATKDYIDFCLLKE---DVNPFISQIELRPLPEEYLH--GFGT 924
Query: 188 FAIGLVARNSFGYSGPSIRYPDDQFDRIWEPFGQSNSTKA-----NTENVSVSGFWNLPP 242
+ L++RN+ G + IR+PDDQ DRIW+ S T A N NV + PP
Sbjct: 925 SVLKLISRNNLGDTNDDIRFPDDQNDRIWKRKETSTPTSALPLSFNVSNVDLKDSVT-PP 1101
Query: 243 SKVFETHLGSEQLESLELRWPTASLPSSKYYIALYFADNT----AGSRIFNISVNGVHYY 298
+V +T L E LE +Y + L+F + AG R+F+I +N
Sbjct: 1102 LQVLQTAL--THPERLEFVHDGLETDDYEYSVFLHFLELNGTVRAGQRVFDIYLNNEIKK 1275
Query: 299 RDLNAIASGV--VVFANQWPLSGPTTITLTPSASSSLGPLINAGEVFNVLSLGGRTSTRD 356
+ +A G A +G ITL ++ S GPL+NA E+ S T+ +D
Sbjct: 1276 EKFDVLAGGSKNSYTALNISANGSLNITLVKASGSEFGPLLNAYEILQARSWIEETNQKD 1455
Query: 357 VIALQRVKESL-----RNPPLD-WSGDPCVPRQYSWTGITCSEGLRIRIVT-LNLTSMDL 409
+ +Q+++E L N L+ WSGDPC+ + W GITC + I+T L+L+S +L
Sbjct: 1456 LEVIQKMREELLLHNQENEALESWSGDPCM--IFPWKGITCDDSTGSSIITKLDLSSNNL 1629
Query: 410 SGSLSSFVANMTALTNIWLGNNSLSGQIPNLSSLTMLETLHLEENQFSGEIPSSLGNISS 469
G++ S V MT L + L +N P+ ++L +L L N SG +P S+ ++
Sbjct: 1630 KGAIPSIVTKMTNLQILNLSHNQFDMLFPSFPPSSLLISLDLSYNDLSGWLPESIISLPH 1809
Query: 470 LKEVFLQNN 478
LK ++ N
Sbjct: 1810 LKSLYFGCN 1836
>NP496346 NP496346|AJ418371.1|CAD10810.1 nod region linked receptor kinase
[Medicago truncatula]
Length = 2706
Score = 145 bits (366), Expect = 3e-35
Identities = 122/402 (30%), Positives = 191/402 (47%), Gaps = 19/402 (4%)
Frame = +1
Query: 68 KTLRSFPLQVKKHCYNIPVYRGAKYMIRTTYFYGGVNGVDHPTPPVFDQIIDGTLWSVVN 127
K +R F + K CYN+P + Y+IR + + +N + + V + G L S
Sbjct: 238 KNVRKFEIYEGKRCYNLPTVKDQVYLIRGIFPFDSLNSSFYVSIGVTEL---GELRSSRL 408
Query: 128 TTVDYANGNSSFYEGVFLAVGKFMSFCIGSNSYTDSDPFVSALEFLILGDSLYNTTDFNN 187
++ EGVF A ++ FC+ D +PF+S +E L + + F
Sbjct: 409 EDLEI--------EGVFRATKDYIDFCLLKE---DVNPFISQIELRPLPEEYLH--GFGT 549
Query: 188 FAIGLVARNSFGYSGPSIRYPDDQFDRIWEPFGQSNSTKA-----NTENVSVSGFWNLPP 242
+ L++RN+ G + IR+PDDQ DRIW+ S T A N NV + PP
Sbjct: 550 SVLKLISRNNLGDTNDDIRFPDDQNDRIWKRKETSTPTSALPLSFNVSNVDLKDSVT-PP 726
Query: 243 SKVFETHLGSEQLESLELRWPTASLPSSKYYIALYFADNT----AGSRIFNISVNGVHYY 298
+V +T L E LE +Y + L+F + AG R+F+I +N
Sbjct: 727 LQVLQTAL--THPERLEFVHDGLETDDYEYSVFLHFLELNGTVRAGQRVFDIYLNNEIKK 900
Query: 299 RDLNAIASGV--VVFANQWPLSGPTTITLTPSASSSLGPLINAGEVFNVLSLGGRTSTRD 356
+ +A G A +G ITL ++ S GPL+NA E+ S T+ +D
Sbjct: 901 EKFDVLAGGSKNSYTALNISANGSLNITLVKASGSEFGPLLNAYEILQARSWIEETNQKD 1080
Query: 357 VIALQRVKESL-----RNPPLD-WSGDPCVPRQYSWTGITCSEGLRIRIVT-LNLTSMDL 409
+ +Q+++E L N L+ WSGDPC+ + W GITC + I+T L+L+S +L
Sbjct: 1081LEVIQKMREELLLHNQENEALESWSGDPCM--IFPWKGITCDDSTGSSIITKLDLSSNNL 1254
Query: 410 SGSLSSFVANMTALTNIWLGNNSLSGQIP-NLSSLTMLETLH 450
G++ S V MT L + L N LSG +P ++ SL L++L+
Sbjct: 1255KGAIPSIVTKMTNLQILDLSYNDLSGWLPESIISLPHLKSLY 1380
>TC87742 weakly similar to GP|7269844|emb|CAB79703.1
serine/threonine-specific receptor protein kinase-like
protein {Arabidopsis thaliana}, partial (29%)
Length = 1318
Score = 114 bits (284), Expect = 1e-25
Identities = 111/390 (28%), Positives = 176/390 (44%), Gaps = 23/390 (5%)
Frame = +1
Query: 2 SILFFLLLFISFHTPSFSQTPPK-GFL-INCGTLT----TTQINNRTWLPDSNFITTGTP 55
SI FFL L + F + GF+ I+CG T + + D +I TG
Sbjct: 163 SIGFFLFLVVCFGVAVLVHAQQQTGFISIDCGGPANFEYTDDVTEIKYGTDGAYIQTGLN 342
Query: 56 KNITTQVLLP-------TLKTLRSFPLQVKKHCYNIPVYR-GAKYMIRTTYFYGGVNGVD 107
+NI+ P T LRSFP Q ++CY + R G+ ++IR ++ YG +G +
Sbjct: 343 RNISANYAYPKNPNLPYTFSDLRSFP-QGNRNCYRLIAGRNGSLHLIRASFLYGNYDGEN 519
Query: 108 HPTPPVFDQIIDGTLWSVVNTTVDYANGNSSF-YEGVFLAVGKFMSFCIGSNSYTDSDPF 166
P FD + LWS TV + N + E + +A + + C+ + PF
Sbjct: 520 KL--PEFDLYVGVNLWS----TVKFKNASEGVTLEIISMATSEETNVCLVNKG--KGIPF 675
Query: 167 VSALEFLILGDSLYNTTDFNNFAIGLVARNSFGYSGPSIRYPDDQFDRIWEPFGQSNSTK 226
+SALE + S+Y T ++ ++ L R G S RY DD +DRIW P S+
Sbjct: 676 ISALELRPVDSSIYKTEFGDSASLLLFKRWDIGSFNGSGRYQDDVYDRIWFPLNSSSWKS 855
Query: 227 ANTEN-VSVSG-FWNLPPSKVFETHLGSEQLESLELRWPTASLPSSKYYIALYFAD---- 280
NT + + VSG + P + + SL+ W + P+ K+ + LYFA+
Sbjct: 856 INTSSKIDVSGDGYKAPFEVISNAATPRNENVSLDFSW-ISDDPNLKFNVYLYFAEFEKL 1032
Query: 281 NTAGSRIFNISVNGVHYYRD-LNAIASGVVVFANQWPLSGPTTITLTPSASSSLGPLINA 339
N R FN+S NG + VF ++ ++ I++ + S+L P++NA
Sbjct: 1033NKTQLRKFNVSWNGSPLINSTIPKYLQSTTVFNSKSLVANEHRISIRKTEDSTLPPILNA 1212
Query: 340 GEVFNVLSLGG-RTSTRDVIALQRVKESLR 368
E++ V TS D A++ VK+S R
Sbjct: 1213VEIYVVRQHDALPTSEEDAEAIRSVKDSYR 1302
>AW688582 similar to GP|11072027|g F12A21.14 {Arabidopsis thaliana}, partial
(11%)
Length = 675
Score = 110 bits (276), Expect = 1e-24
Identities = 58/161 (36%), Positives = 95/161 (58%), Gaps = 5/161 (3%)
Frame = +1
Query: 331 SSLGPLINAGEVFNVLSLGGRTSTRD---VIALQRVKESL--RNPPLDWSGDPCVPRQYS 385
S+ GPL+NA E+ + +T +D V A + + + +N GDPCVP +
Sbjct: 175 STRGPLLNAMEISKYQEIASKTFKQDSNFVNAFSSLSDEIIPKN-----EGDPCVPTPWE 339
Query: 386 WTGITCSEGLRIRIVTLNLTSMDLSGSLSSFVANMTALTNIWLGNNSLSGQIPNLSSLTM 445
W + CS RI +NL+ +L+G + + NM ALT +WL N L+GQ+P++S+L
Sbjct: 340 W--VNCSTATPARITNINLSGRNLTGEIPRELNNMEALTELWLXRNLLTGQLPDMSNLIN 513
Query: 446 LETLHLEENQFSGEIPSSLGNISSLKEVFLQNNNLTGQIPA 486
L+ +HLE N+ +G +P+ LG++ L+ + +QNN+ TG IP+
Sbjct: 514 LKIMHLENNKLTGPLPTYLGSLPGLQALXIQNNSFTGDIPS 636
>BF638005 weakly similar to GP|6560758|gb| F3M18.23 {Arabidopsis thaliana},
partial (31%)
Length = 672
Score = 105 bits (262), Expect = 4e-23
Identities = 70/180 (38%), Positives = 101/180 (55%), Gaps = 7/180 (3%)
Frame = +3
Query: 316 PLSGPT-TITLTPSASSSLGPLINAGEVFNVLSLGGRTSTRDVIALQRVKESLRNPP-LD 373
P++G + ITL P S I A E+ V+ +T + +V+ALQR+K+ L PP
Sbjct: 48 PVNGRSLAITLRPKEGSLA--TITAIEILEVIVPESKTLSDEVMALQRLKKDLGLPPRFG 221
Query: 374 WSGDPCVPRQYSWTGITC----SEGLRIRIVTLNLTSMDLSGSLSSFVANMTALTNIWLG 429
W+GDPCVP+Q+ W G+ C S G + I L L + L G L ++ + L I L
Sbjct: 222 WNGDPCVPQQHPWIGVDCQLDKSSGNWV-IDGLGLDNQGLKGFLPKDISRLHNLQIINLS 398
Query: 430 NNSLSGQIPN-LSSLTMLETLHLEENQFSGEIPSSLGNISSLKEVFLQNNNLTGQIPANL 488
NS+ G IP+ L ++T L+ L L N F+G IP SLG ++SLK + L L+G +PA L
Sbjct: 399 GNSIGGAIPSSLGTVTTLQVLDLSYNVFNGSIPDSLGQLTSLKRLNLNGXFLSGMVPATL 578
>CB891120 similar to GP|17979065|gb putative receptor kinase homolog
{Arabidopsis thaliana}, partial (32%)
Length = 845
Score = 98.6 bits (244), Expect = 5e-21
Identities = 60/140 (42%), Positives = 86/140 (60%), Gaps = 7/140 (5%)
Frame = +2
Query: 356 DVIALQRVKESLRNPPL------DWSGDPCVPRQYSWTGITCSEGLRIRIVTLNLTSMDL 409
+V+AL +KE+L +P ++S DPC SW ITCS + + L S L
Sbjct: 116 EVVALMSIKEALNDPHNVLSNWDEFSVDPC-----SWAMITCSSDSFV--IGLGAPSQSL 274
Query: 410 SGSLSSFVANMTALTNIWLGNNSLSGQIP-NLSSLTMLETLHLEENQFSGEIPSSLGNIS 468
SG+LSS +AN+T L + L NN++SG+IP L +L L+TL L N+FSG IPSSL ++
Sbjct: 275 SGTLSSSIANLTNLKQVLLQNNNISGKIPPELGNLPKLQTLDLSNNRFSGFIPSSLNQLN 454
Query: 469 SLKEVFLQNNNLTGQIPANL 488
SL+ + L NN+L+G P +L
Sbjct: 455 SLQYMRLNNNSLSGPFPVSL 514
>TC81839 similar to GP|21593619|gb|AAM65586.1 receptor protein kinase-like
protein {Arabidopsis thaliana}, partial (34%)
Length = 960
Score = 97.4 bits (241), Expect = 1e-20
Identities = 59/138 (42%), Positives = 89/138 (63%), Gaps = 5/138 (3%)
Frame = +2
Query: 356 DVIALQRVKESLRNPP---LDWSGDPCVPRQYSWTGITCS-EGLRIRIVTLNLTSMDLSG 411
+V AL +KESL +P +W GD P SW +TCS E L +V+L + S +LSG
Sbjct: 230 EVQALVSIKESLMDPHGIFENWDGDAVDP--CSWNMVTCSPENL---VVSLGIPSQNLSG 394
Query: 412 SLSSFVANMTALTNIWLGNNSLSGQIPN-LSSLTMLETLHLEENQFSGEIPSSLGNISSL 470
+LSS + N+T L + L NN+++G IP+ L L+ML+TL L +N F G+IP SLG++ +L
Sbjct: 395 TLSSSIGNLTNLQTVVLQNNNITGPIPSELGKLSMLQTLDLSDNLFHGKIPPSLGHLRNL 574
Query: 471 KEVFLQNNNLTGQIPANL 488
+ + L NN+ +G+ P +L
Sbjct: 575 QYLRLNNNSFSGECPESL 628
>TC78630 similar to GP|10177052|dbj|BAB10464. receptor-like protein kinase
{Arabidopsis thaliana}, partial (43%)
Length = 1445
Score = 90.9 bits (224), Expect = 1e-18
Identities = 52/120 (43%), Positives = 72/120 (59%), Gaps = 4/120 (3%)
Frame = +3
Query: 373 DWSG---DPCVPRQYSWTGITCSEGLRIRIVTLNLTSMDLSGSLSSFVANMTALTNIWLG 429
DW PC +SW+ +TC G +++L L S+ SG+LS + + L N+ L
Sbjct: 282 DWDSHLVSPC----FSWSHVTCRNG---HVISLTLASIGFSGTLSPSITRLKYLVNLELQ 440
Query: 430 NNSLSGQIPN-LSSLTMLETLHLEENQFSGEIPSSLGNISSLKEVFLQNNNLTGQIPANL 488
NN+LSG IP+ +S+LT L+ L+L N F+G IP S G +SSLK V L +N LTG IP L
Sbjct: 441 NNNLSGPIPDYISNLTDLQYLNLANNNFNGSIPVSWGQLSSLKNVDLSSNGLTGTIPTQL 620
>TC91051 similar to PIR|H84787|H84787 probable receptor-like protein kinase
[imported] - Arabidopsis thaliana, partial (36%)
Length = 1240
Score = 56.2 bits (134), Expect(2) = 1e-18
Identities = 72/250 (28%), Positives = 105/250 (41%), Gaps = 29/250 (11%)
Frame = +3
Query: 165 PFVSALEFLILGDSLYNTTDFNNFAIGLVARNSFGYSGPS-IRYPDDQFDRIWEPFGQSN 223
PF+S LE S+Y T F + + AR +FG + IRYPDD FDRIWE +
Sbjct: 516 PFISTLELRQFNGSIYYTQFEQQFYLSVSARINFGAETDAPIRYPDDPFDRIWE---SDS 686
Query: 224 STKAN--------TENVSVS-----GFWNLPPSKVFETHLGSEQLESLELRWPTASLPSS 270
KAN TE VS + ++PP KV +T + SL R P +
Sbjct: 687 VKKANYLVDVAVGTEKVSTNVPILVNRDDVPPVKVMQTAVVGTN-GSLTYRLNLDGFPGN 863
Query: 271 KYYIALYFAD----NTAGSRIFNISVNGVHYYR------DLNAIASGVVVFANQWPLSGP 320
+ + YFA+ + SR F + + G + NA + LS P
Sbjct: 864 AWAVT-YFAEIEDLSPNESRKFRLVLPGQPEISKAIVNIEENAFGKYRLYEPGFTNLSLP 1040
Query: 321 TTIT--LTPSASSSLGPLINAGEVFNVLSLGGRTSTRDVIALQRVKESLRNPPLDWS--- 375
++ + SS GPL+NA E+ L + DV A+ V + +W+
Sbjct: 1041FVLSFKFAKTPDSSKGPLVNAMEINKYLE--KNDGSPDVEAISGVLSHYSS--ANWTQER 1208
Query: 376 GDPCVPRQYS 385
DPC+P +S
Sbjct: 1209SDPCLPVPWS 1238
Score = 54.7 bits (130), Expect(2) = 1e-18
Identities = 54/178 (30%), Positives = 86/178 (47%), Gaps = 10/178 (5%)
Frame = +2
Query: 2 SILFFLL-----LFISFHTPSFSQTPPKGFL-INCG-TLTTTQINNRTWLPDSNFITTGT 54
++LFFL L + P S +GF+ ++CG + + T W PD N +T G
Sbjct: 26 NLLFFLFFPSLPLSLFLLLPFSSSQIMQGFVSLDCGGSESFTDDIGLDWTPD-NKLTYGE 202
Query: 55 PKNITTQVLLPTLK---TLRSFPLQVKKHCYNIPVYRGAKYMIRTTYFYGGVNGVDHPTP 111
I+ V+ T K TLR FP +K+CY + V +Y++R ++ YG N ++
Sbjct: 203 ISTIS--VVNETRKQYTTLRHFPADSRKYCYTLDVISRTRYLLRASFLYG--NFDNNNVY 370
Query: 112 PVFDQIIDGTLWSVVNTTVDYANGNSSFYEGVFLAVGKFMSFCIGSNSYTDSDPFVSA 169
P FD + T WS + + AN E +FLA +S C+ SN+ T + ++ A
Sbjct: 371 PKFDISVGATHWSTI--VISDAN-IIEMRELIFLASSSTVSVCL-SNATTGTAVYIYA 532
>TC82585 similar to GP|7715608|gb|AAF68126.1| F20B17.5 {Arabidopsis
thaliana}, partial (35%)
Length = 1288
Score = 86.7 bits (213), Expect = 2e-17
Identities = 56/143 (39%), Positives = 78/143 (54%), Gaps = 4/143 (2%)
Frame = +3
Query: 352 TSTRDVIALQRVKESLRNPPLDW--SGDPCVPRQYSWTGITCSEGLRIRIVTLNLTSMDL 409
T +DV AL+ +K+ N P W S DPC W G+TC++ R+ +L L++M L
Sbjct: 192 TDPQDVAALRSLKDIWENTPPSWDKSDDPCGA---PWEGVTCNKS---RVTSLGLSTMGL 353
Query: 410 SGSLSSFVANMTALTNIWLG-NNSLSGQI-PNLSSLTMLETLHLEENQFSGEIPSSLGNI 467
G LS + +T L ++ L N L G I P L L+ L L L FSG IP LG++
Sbjct: 354 KGKLSGDIGGLTELRSLDLSFNKDLMGPISPELGDLSKLNILILAGCSFSGNIPDKLGDL 533
Query: 468 SSLKEVFLQNNNLTGQIPANLLK 490
S L + L +NN TG+IP +L K
Sbjct: 534 SELSFLALNSNNFTGKIPPSLGK 602
Score = 53.9 bits (128), Expect = 1e-07
Identities = 37/107 (34%), Positives = 57/107 (52%), Gaps = 3/107 (2%)
Frame = +3
Query: 387 TGITCSEGLRIRIVTLNLTSMDLSGSLSS--FVANMTALTNIWLGNNSLSGQIPNLSSLT 444
T T L ++ + LSGS+ F ++M L +I N LSG IP+ L
Sbjct: 666 TSTTPGLDLLLKAKHFHFNKNQLSGSIPPQLFSSDMV-LIHILFDRNDLSGSIPSTIGLV 842
Query: 445 M-LETLHLEENQFSGEIPSSLGNISSLKEVFLQNNNLTGQIPANLLK 490
+E L L+ N +GE+PS+L + ++ E+ L +NNL+G +P NL K
Sbjct: 843 QTVEVLRLDRNFLTGEVPSNLNKLGNINELNLAHNNLSGSLP-NLTK 980
Score = 47.8 bits (112), Expect = 1e-05
Identities = 37/117 (31%), Positives = 58/117 (48%), Gaps = 9/117 (7%)
Frame = +3
Query: 385 SWTG-ITCSEGLRIRIVTLNLTSMDLSGSLSSFVANMTALT------NIWLGNNSLSGQI 437
++TG I S G ++ L+L L+G L + L + N LSG I
Sbjct: 567 NFTGKIPPSLGKLSKLYWLDLADNQLTGPLPVSTSTTPGLDLLLKAKHFHFNKNQLSGSI 746
Query: 438 PN--LSSLTMLETLHLEENQFSGEIPSSLGNISSLKEVFLQNNNLTGQIPANLLKPG 492
P SS +L + + N SG IPS++G + +++ + L N LTG++P+NL K G
Sbjct: 747 PPQLFSSDMVLIHILFDRNDLSGSIPSTIGLVQTVEVLRLDRNFLTGEVPSNLNKLG 917
Score = 34.7 bits (78), Expect = 0.087
Identities = 19/63 (30%), Positives = 33/63 (52%)
Frame = +3
Query: 394 GLRIRIVTLNLTSMDLSGSLSSFVANMTALTNIWLGNNSLSGQIPNLSSLTMLETLHLEE 453
GL + L L L+G + S + + + + L +N+LSG +PNL+ +T L + L +
Sbjct: 834 GLVQTVEVLRLDRNFLTGEVPSNLNKLGNINELNLAHNNLSGSLPNLTKMTSLNYVDLRQ 1013
Query: 454 NQF 456
F
Sbjct: 1014QLF 1022
>BE204732 weakly similar to GP|8978275|db receptor protein kinase-like
{Arabidopsis thaliana}, partial (10%)
Length = 461
Score = 85.5 bits (210), Expect = 4e-17
Identities = 52/142 (36%), Positives = 81/142 (56%), Gaps = 4/142 (2%)
Frame = +1
Query: 351 RTSTRDVIALQRVKESLRNPPLDW--SGDPCVPRQYSWTGITCSEGLRIRIVTLNLTSMD 408
+TS D +AL +K+ +N P W S DPC W GI CS R++T++L+SMD
Sbjct: 46 KTSNDDYLALSTLKDEWKNVPPSWEDSEDPCGDH---WEGIECSNS---RVITISLSSMD 207
Query: 409 LSGSLSSFVANMTALTNIWLG-NNSLSGQIP-NLSSLTMLETLHLEENQFSGEIPSSLGN 466
LSG LSS + +++ L + L N L+G +P + +L L L L F+G IP ++GN
Sbjct: 208 LSGQLSSEIGSLSELQILVLSYNKDLTGPLPAEIGNLKKLTNLQLINCGFTGPIPDTIGN 387
Query: 467 ISSLKEVFLQNNNLTGQIPANL 488
+ L + L +N +G+IP ++
Sbjct: 388 LQRLVFLSLNSNRFSGRIPPSI 453
>TC82234 weakly similar to GP|7715608|gb|AAF68126.1| F20B17.5 {Arabidopsis
thaliana}, partial (12%)
Length = 760
Score = 84.7 bits (208), Expect = 7e-17
Identities = 62/205 (30%), Positives = 97/205 (47%), Gaps = 3/205 (1%)
Frame = +2
Query: 287 IFNISVNGVHYYRDLNAIASGVVVFANQWPLSGPTTITLTPSASSSLGPLINAGEVFNVL 346
+F ++N +H+ N+I+ V + P + L +F VL
Sbjct: 83 LF*FNINSIHFES*FNSISEQVFCVMEEQHKVFPLLLLL----------------LFQVL 214
Query: 347 SLGGRTSTRDVIALQRVKESLRNPPLDWSG-DPCVPRQYSWTGITCSEGLRIRIVTLNLT 405
+ +T D AL + +S N P +W G DPC +W GI C RI L L
Sbjct: 215 HVASQTDRGDFTALSSLTQSWNNRPSNWVGSDPCGS---NWAGIGCDNS---RITELKLL 376
Query: 406 SMDLSGSLSSFVANMTALTNIWLGNNS-LSGQIPN-LSSLTMLETLHLEENQFSGEIPSS 463
+ L G LSS + +++ L + L +N+ ++G IP + +L L +L L FSG IP S
Sbjct: 377 GLSLEGQLSSAIQSLSELETLDLSSNTGMTGTIPREIGNLKNLNSLALVGCGFSGPIPDS 556
Query: 464 LGNISSLKEVFLQNNNLTGQIPANL 488
+G++ L + L +NN TG IP +L
Sbjct: 557 IGSLKKLTFLALNSNNFTGNIPHSL 631
>AW256777 weakly similar to PIR|T01269|T01 serine/threonine-specific protein
kinase (EC 2.7.1.-) F27F23.1 - Arabidopsis thaliana,
partial (13%)
Length = 373
Score = 81.6 bits (200), Expect = 6e-16
Identities = 44/118 (37%), Positives = 70/118 (59%), Gaps = 2/118 (1%)
Frame = +3
Query: 324 TLTPSASSSLGPLINAGEVFNVLSLGGRTSTRDVIALQRVKESLRNPPLDWSGDPCVPRQ 383
TL+ + +S+L P++NA EV+ V + + +D + R + +W GDPC P
Sbjct: 18 TLSKTDNSTLPPILNAVEVYKVKNFSQSETQQDDVDTMRNIKKAYGVARNWQGDPCGPVN 197
Query: 384 YSWTGITCS-EGLRI-RIVTLNLTSMDLSGSLSSFVANMTALTNIWLGNNSLSGQIPN 439
Y W G+ CS +G I RI +LNL+S L+G +SS ++ +T L + L NNSL+G +P+
Sbjct: 198 YMWEGLNCSLDGNNIPRITSLNLSSSGLTGEISSSISKLTMLQYLDLSNNSLNGSLPD 371
>TC80585 weakly similar to GP|9759164|dbj|BAB09720.1 receptor kinase-like
protein {Arabidopsis thaliana}, partial (24%)
Length = 900
Score = 81.6 bits (200), Expect = 6e-16
Identities = 50/143 (34%), Positives = 81/143 (55%), Gaps = 8/143 (5%)
Frame = +3
Query: 354 TRDVIALQRVKESLRNPP----LDWS---GDPCVPRQYSWTGITCSEGLRIRIVTLNLTS 406
+ +V AL KE++ P +W+ DPC W G++C+ G R ++ LN++
Sbjct: 414 SNEVGALSTFKEAVYEDPHMVLSNWNTLDSDPC-----DWNGVSCT-GTRDHVIKLNISG 575
Query: 407 MDLSGSLSSFVANMTALTNIWLGNNSLSGQIPN-LSSLTMLETLHLEENQFSGEIPSSLG 465
L G L+ + +T L + L N+L G IP LS+L L+ L L +NQ SG IP +G
Sbjct: 576 ALLRGFLAPEIGKITYLQELILHGNNLIGTIPKELSALKSLKVLDLGKNQLSGPIPREIG 755
Query: 466 NISSLKEVFLQNNNLTGQIPANL 488
N++ + ++ LQ+N LTG++P+ L
Sbjct: 756 NLTQIVKINLQSNGLTGRLPSEL 824
>BG451633 weakly similar to GP|16930691|gb AT4g26540/M3E9_30 {Arabidopsis
thaliana}, partial (7%)
Length = 639
Score = 81.3 bits (199), Expect = 8e-16
Identities = 54/154 (35%), Positives = 81/154 (52%), Gaps = 7/154 (4%)
Frame = +1
Query: 342 VFNVLSLGGRTSTRDVIALQRVKESLRNPPLD----WSGDPCVPRQYS--WTGITCSEGL 395
+F L++ +T T D AL K S+ P D WS S W G+TC E
Sbjct: 64 LFTCLAMISKTITTDEFALLAFKSSITLDPYDILSNWSISSSTSSFSSCNWVGVTCDEH- 240
Query: 396 RIRIVTLNLTSMDLSGSLSSFVANMTALTNIWLGNNSLSGQIPN-LSSLTMLETLHLEEN 454
R+ L+L +MDL G++S + N++ L + L NS G++P+ L L L+ L L +N
Sbjct: 241 HGRVNALDLNNMDLEGTISPQLGNLSFLVVLDLQGNSFYGELPHELLQLKRLKWLDLSDN 420
Query: 455 QFSGEIPSSLGNISSLKEVFLQNNNLTGQIPANL 488
F GEIPS +G+++ L + L NN G IP ++
Sbjct: 421 DFVGEIPSRIGDLAKLHHLDLYFNNFVGAIPQSI 522
>TC89894 weakly similar to GP|14495542|gb|AAB36558.2 receptor-like protein
kinase INRPK1 {Ipomoea nil}, partial (7%)
Length = 780
Score = 79.0 bits (193), Expect = 4e-15
Identities = 45/118 (38%), Positives = 67/118 (56%), Gaps = 1/118 (0%)
Frame = +3
Query: 374 WSGDPCVPRQYSWTGITCSEGLRIRIVTLNLTSMDLSGSLSSFVANMTALTNIWLGNNSL 433
W+ P SW G+ CS+ + + +L+L+ +SG L + + L + L N L
Sbjct: 345 WNSSHSTP--CSWKGVECSDD-SLNVTSLSLSDHSISGQLGPEIGKLIHLQLLDLSINDL 515
Query: 434 SGQIP-NLSSLTMLETLHLEENQFSGEIPSSLGNISSLKEVFLQNNNLTGQIPANLLK 490
SG+IP LS+ ML+ L L EN FSGEIPS L N S L+ ++L N+ G+IP +L +
Sbjct: 516 SGEIPIELSNCNMLQYLDLSENNFSGEIPSELSNCSMLQYLYLSVNSFRGEIPQSLFQ 689
Score = 75.1 bits (183), Expect = 6e-14
Identities = 48/138 (34%), Positives = 72/138 (51%), Gaps = 12/138 (8%)
Frame = +3
Query: 366 SLRNPPLDWSGDPCVPRQYSWTGITCSE-----------GLRIRIVTLNLTSMDLSGSLS 414
S + P W G C + T ++ S+ G I + L+L+ DLSG +
Sbjct: 351 SSHSTPCSWKGVECSDDSLNVTSLSLSDHSISGQLGPEIGKLIHLQLLDLSINDLSGEIP 530
Query: 415 SFVANMTALTNIWLGNNSLSGQIPN-LSSLTMLETLHLEENQFSGEIPSSLGNISSLKEV 473
++N L + L N+ SG+IP+ LS+ +ML+ L+L N F GEIP SL I+ L+++
Sbjct: 531 IELSNCNMLQYLDLSENNFSGEIPSELSNCSMLQYLYLSVNSFRGEIPQSLFQINPLEDL 710
Query: 474 FLQNNNLTGQIPANLLKP 491
L NN+L G IP KP
Sbjct: 711 RLNNNSLNGSIPRWYWKP 764
>TC79788 weakly similar to GP|21554189|gb|AAM63268.1 putative leucine-rich
repeat disease resistance protein {Arabidopsis
thaliana}, partial (52%)
Length = 1484
Score = 77.0 bits (188), Expect = 2e-14
Identities = 55/150 (36%), Positives = 84/150 (55%), Gaps = 13/150 (8%)
Frame = +3
Query: 352 TSTRDVIALQRVKESLRNPPLD-WSG--------DPCVP--RQYSWTGITCS-EGLRIRI 399
TS D+ AL+ K S++ + WS DPC R Y G TC+ + I
Sbjct: 150 TSPSDIEALKAFKASIKPSSITPWSCVASWNFTIDPCSHPRRTYFLCGFTCTTDSTHINQ 329
Query: 400 VTLNLTSMDLSGSLSSFVANMTALTNIWLGNNSLSGQIPN-LSSLTMLETLHLEENQFSG 458
+TL+ S SGSL+ ++ +T L + L +N+ G IP+ +SSL+ L+TL L N FSG
Sbjct: 330 ITLDPASY--SGSLTPLISKLTQLITLDLSDNNFFGSIPSSISSLSNLKTLTLRSNSFSG 503
Query: 459 EIPSSLGNISSLKEVFLQNNNLTGQIPANL 488
IP S+ ++ SL+ + L +N+LTG +P +L
Sbjct: 504 PIPPSIISLKSLESLDLSHNSLTGSLPNSL 593
Score = 40.4 bits (93), Expect = 0.002
Identities = 33/105 (31%), Positives = 51/105 (48%), Gaps = 23/105 (21%)
Frame = +3
Query: 401 TLNLTSMDLSGSLSSFVANMTALTNIWLGNNSLSGQIPNL-----------SSLT----- 444
+L+L+ L+GSL + + ++ L I L N L+G IP L +SL+
Sbjct: 543 SLDLSHNSLTGSLPNSLNSLINLHRIDLSFNKLAGSIPKLPPNLLELAIKANSLSGPLQK 722
Query: 445 -------MLETLHLEENQFSGEIPSSLGNISSLKEVFLQNNNLTG 482
LE + L EN +G I + +SSL++V L NN+ TG
Sbjct: 723 TTFEGSNQLEVVELSENALTGTIETWFLLLSSLQQVNLANNSFTG 857
Score = 38.1 bits (87), Expect = 0.008
Identities = 59/253 (23%), Positives = 101/253 (39%), Gaps = 10/253 (3%)
Frame = +3
Query: 222 SNSTKANTENVSVSGFWNLPPSKVFETHLGSEQLESLELRWPTASLPSSKYYIALYFADN 281
S T+ T ++S + F+ PS + L++L LR + S P I+L ++
Sbjct: 378 SKLTQLITLDLSDNNFFGSIPSSISSL----SNLKTLTLRSNSFSGPIPPSIISLKSLES 545
Query: 282 T--AGSRIFNISVNGVHYYRDLNAIASGVVVFANQWPLSGPTTITLTPSASSSLGPL--- 336
+ + + N ++ +L+ I A P P + L A+S GPL
Sbjct: 546 LDLSHNSLTGSLPNSLNSLINLHRIDLSFNKLAGSIPKLPPNLLELAIKANSLSGPLQKT 725
Query: 337 -INAGEVFNVLSLGGRTSTRDVIALQRVKESLRNPPLDWSGDPCVPRQYSWTGITCSE-- 393
V+ L T + + SL+ L S+TGI S+
Sbjct: 726 TFEGSNQLEVVELSENALTGTIETWFLLLSSLQQVNL---------ANNSFTGIQISKPA 878
Query: 394 -GLRIRIVTLNLTSMDLSGSLSSFVANMTALTNIWLGNNSLSGQIP-NLSSLTMLETLHL 451
G+ +V LNL + G + +A L+ + + +NSL G IP + ++ L L
Sbjct: 879 RGVESNLVALNLGFNRIQGYAPANLAAYPLLSFLSIRHNSLRGNIPLEYGQIKSMKRLFL 1058
Query: 452 EENQFSGEIPSSL 464
+ N F G+ P +L
Sbjct: 1059DGNFFDGKPPPAL 1097
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.318 0.136 0.409
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 16,932,129
Number of Sequences: 36976
Number of extensions: 251021
Number of successful extensions: 2006
Number of sequences better than 10.0: 248
Number of HSP's better than 10.0 without gapping: 1519
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1815
length of query: 496
length of database: 9,014,727
effective HSP length: 100
effective length of query: 396
effective length of database: 5,317,127
effective search space: 2105582292
effective search space used: 2105582292
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 60 (27.7 bits)
Medicago: description of AC137838.4


