

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137838.13 + phase: 0 /pseudo
(1485 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
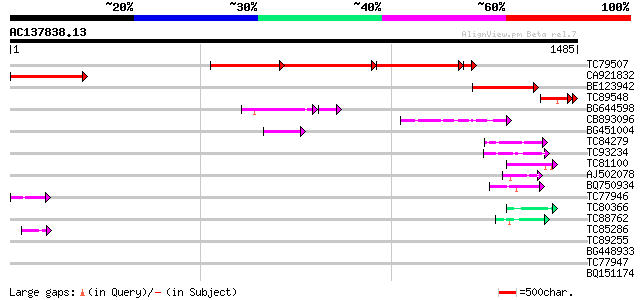
Score E
Sequences producing significant alignments: (bits) Value
TC79507 similar to GP|14194103|gb|AAK56246.1 AT3g22390/MCB17_12 ... 500 0.0
CA921832 similar to GP|14194103|gb AT3g22390/MCB17_12 {Arabidops... 417 e-116
BE123942 similar to GP|14194103|gb AT3g22390/MCB17_12 {Arabidops... 329 6e-90
TC89548 similar to GP|7300093|gb|AAF55262.1| CG10278 gene produc... 155 1e-40
BG644598 weakly similar to PIR|T48107|T481 hypothetical protein ... 95 4e-23
CB893096 80 8e-15
BG451004 similar to GP|14194103|gb AT3g22390/MCB17_12 {Arabidops... 50 7e-06
TC84279 weakly similar to GP|1799947|dbj|BAA16433.1 BENZENE 1 2-... 49 1e-05
TC93234 weakly similar to GP|21751020|dbj|BAC03887. unnamed prot... 47 7e-05
TC81100 similar to PIR|H84824|H84824 En/Spm-like transposon prot... 47 7e-05
AJ502078 similar to GP|15529165|gb AT3g04880/T9J14_17 {Arabidops... 46 9e-05
BQ750934 similar to PIR|T19673|T19 hypothetical protein C33B4.3 ... 45 2e-04
TC77946 homologue to GP|10334491|emb|CAC10207. putative splicing... 44 5e-04
TC80366 similar to GP|13877617|gb|AAK43886.1 protein kinase-like... 44 5e-04
TC88762 similar to GP|22136928|gb|AAM91808.1 unknown protein {Ar... 43 8e-04
TC85286 similar to GP|3776572|gb|AAC64889.1| ESTs gb|R65052 gb|... 43 8e-04
TC89255 similar to GP|3135257|gb|AAC16457.1| unknown protein {Ar... 42 0.002
BG448933 similar to PIR|T09648|T09 nucleolin homolog nuM1 - alfa... 42 0.002
TC77947 homologue to GP|10334491|emb|CAC10207. putative splicing... 42 0.002
BQ151174 similar to GP|11036868|gb| PxORF73 peptide {Plutella xy... 40 0.009
>TC79507 similar to GP|14194103|gb|AAK56246.1 AT3g22390/MCB17_12
{Arabidopsis thaliana}, partial (11%)
Length = 2117
Score = 500 bits (1288), Expect(4) = 0.0
Identities = 245/251 (97%), Positives = 247/251 (97%)
Frame = +3
Query: 710 PGSNLSVVPVPSTELHSGNIHGRATNSTQDKGPSLAMFPGHIGKDKSSQPSNVDNSSRKP 769
P + L + PVPSTELHSGNIHGRATNSTQDKGPSLAMFPGHIGKDKSSQPSNVDNSSRKP
Sbjct: 555 PAAILVLYPVPSTELHSGNIHGRATNSTQDKGPSLAMFPGHIGKDKSSQPSNVDNSSRKP 734
Query: 770 ILLQQTLPSGAAPSNILHGPTFIFPLNQQQAAAAAAAASVRPGSVKSLPVTSNGPPSSTT 829
ILLQQTLPSGAAPSNILHGPTFIFPLNQQQAAAAAAAASVRPGSVKSLPVTSNGPPSSTT
Sbjct: 735 ILLQQTLPSGAAPSNILHGPTFIFPLNQQQAAAAAAAASVRPGSVKSLPVTSNGPPSSTT 914
Query: 830 NSAPPNTSGAGAAAPAPPTMSFTYPNMSGNETQYMAILQNNAYPFPIPAHVGGPPGYRGN 889
NSAPPNTSGAGAAAPAPPTMSFTYPNMSGNETQYMAILQNNAYPFPIPAHVGGPPGYRGN
Sbjct: 915 NSAPPNTSGAGAAAPAPPTMSFTYPNMSGNETQYMAILQNNAYPFPIPAHVGGPPGYRGN 1094
Query: 890 PAQAFPFFNGSFYPSQMIHPSQIQSQQPPAQSQQSQQGHPNTTISTGSSSSQKHAQNQQQ 949
PAQAFPFFNGSFYPSQMIHPSQIQSQQPPAQSQQSQQGHPNTTISTGSSSSQKHAQNQQQ
Sbjct: 1095PAQAFPFFNGSFYPSQMIHPSQIQSQQPPAQSQQSQQGHPNTTISTGSSSSQKHAQNQQQ 1274
Query: 950 KANNASGSNGG 960
KANNASGSNGG
Sbjct: 1275KANNASGSNGG 1307
Score = 457 bits (1177), Expect(4) = 0.0
Identities = 229/229 (100%), Positives = 229/229 (100%)
Frame = +2
Query: 961 GSGSLQGFPVTKNPQSQILQLQQQQHRQQLHNHHTSNAARQVESEMGCEDSPSTADSRHN 1020
GSGSLQGFPVTKNPQSQILQLQQQQHRQQLHNHHTSNAARQVESEMGCEDSPSTADSRHN
Sbjct: 1310 GSGSLQGFPVTKNPQSQILQLQQQQHRQQLHNHHTSNAARQVESEMGCEDSPSTADSRHN 1489
Query: 1021 RATMNIYGQNFAMPMQTPNFALMTTAMSGAGSNGNHSEKKQQQQHPGSKAGGETSPAFAM 1080
RATMNIYGQNFAMPMQTPNFALMTTAMSGAGSNGNHSEKKQQQQHPGSKAGGETSPAFAM
Sbjct: 1490 RATMNIYGQNFAMPMQTPNFALMTTAMSGAGSNGNHSEKKQQQQHPGSKAGGETSPAFAM 1669
Query: 1081 PFPSITGATALDLSSIAQNHSIMQSSHNFQLMATAQAASAQLKKSYHAAEEGKHVVNSSN 1140
PFPSITGATALDLSSIAQNHSIMQSSHNFQLMATAQAASAQLKKSYHAAEEGKHVVNSSN
Sbjct: 1670 PFPSITGATALDLSSIAQNHSIMQSSHNFQLMATAQAASAQLKKSYHAAEEGKHVVNSSN 1849
Query: 1141 LEEDRKAISGKIPTTGGQNIAFARSDVADPSMTSIACNNVIDSSGRSLN 1189
LEEDRKAISGKIPTTGGQNIAFARSDVADPSMTSIACNNVIDSSGRSLN
Sbjct: 1850 LEEDRKAISGKIPTTGGQNIAFARSDVADPSMTSIACNNVIDSSGRSLN 1996
Score = 398 bits (1022), Expect(4) = 0.0
Identities = 192/193 (99%), Positives = 192/193 (99%)
Frame = +2
Query: 526 HVMKEDQKSHKMDEVMVVEIEKVKAKAEENEFQRAPIVQKERGIDLQLELEKTDRVDSNG 585
HVMKEDQKSHK DEVMVVEIEKVKAKAEENEFQRAPIVQKERGIDLQLELEKTDRVDSNG
Sbjct: 2 HVMKEDQKSHKRDEVMVVEIEKVKAKAEENEFQRAPIVQKERGIDLQLELEKTDRVDSNG 181
Query: 586 NGNHLNKKQHQNVQRHHHQLQQQTNLEKNVQSNSLPIPMNVPSWPGGLPSMGYMTPLQGV 645
NGNHLNKKQHQNVQRHHHQLQQQTNLEKNVQSNSLPIPMNVPSWPGGLPSMGYMTPLQGV
Sbjct: 182 NGNHLNKKQHQNVQRHHHQLQQQTNLEKNVQSNSLPIPMNVPSWPGGLPSMGYMTPLQGV 361
Query: 646 VSMDGTTMPSAAIPPPHLLFNQPRPKRCATHCHIAQKILYNQQIARMNPFWPAAAGSASL 705
VSMDGTTMPSAAIPPPHLLFNQPRPKRCATHCHIAQKILYNQQIARMNPFWPAAAGSASL
Sbjct: 362 VSMDGTTMPSAAIPPPHLLFNQPRPKRCATHCHIAQKILYNQQIARMNPFWPAAAGSASL 541
Query: 706 YGAKPGSNLSVVP 718
YGAKPGSNLSVVP
Sbjct: 542 YGAKPGSNLSVVP 580
Score = 56.2 bits (134), Expect(4) = 0.0
Identities = 28/32 (87%), Positives = 28/32 (87%)
Frame = +3
Query: 1190 LGSASSRASASGMPSAINSNAAGSQQQMQRNQ 1221
LGSASSRASA GMP AIN NAAGS QQMQRNQ
Sbjct: 1998 LGSASSRASAXGMPXAINXNAAGSXQQMQRNQ 2093
Score = 34.7 bits (78), Expect = 0.28
Identities = 45/205 (21%), Positives = 75/205 (35%), Gaps = 35/205 (17%)
Frame = +3
Query: 1230 NQFAAAAVAAARNKTPSTSNGSIYSDNLPSTSSISTKFPN-----AVSAFPQSLVQSSNT 1284
NQ AAA AAA + P + + N P +S+ ++ PN A + P ++ +
Sbjct: 813 NQQQAAAAAAAASVRPGSVKSLPVTSNGPPSSTTNSAPPNTSGAGAAAPAPPTMSFTYPN 992
Query: 1285 VVTQSSQW----KNSA-----------------------RVTNTSLSPQTMASPPSSSVK 1317
+ +Q+ +N+A N S P M P +
Sbjct: 993 MSGNETQYMAILQNNAYPFPIPAHVGGPPGYRGNPAQAFPFFNGSFYPSQMIHPSQIQSQ 1172
Query: 1318 NPPQQQARSQQGHTQISFAANPKSSTPQVQT---ASSTQSPSPPVMVGSPTNSSMSKNTG 1374
PP Q +SQQGH + + SS Q ++ S S V +S+ T
Sbjct: 1173 QPPAQSQQSQQGHPNTTISTGSSSSQKHAQNQQQKANNASGSNGGWVVEAYKVFLSQKTR 1352
Query: 1375 SPRTTNSTSTNNKTSQTSSLSSQQP 1399
+ R + + + +S T + QP
Sbjct: 1353 NRRFYSCNNNSTGSSCTITTLQTQP 1427
>CA921832 similar to GP|14194103|gb AT3g22390/MCB17_12 {Arabidopsis
thaliana}, partial (3%)
Length = 802
Score = 417 bits (1072), Expect = e-116
Identities = 202/202 (100%), Positives = 202/202 (100%)
Frame = -1
Query: 1 MDRIREARRSTMAANGLTRRRHRTNSLRDSPDDDGGMEMQEPTRLRDRGSGKKERDRERE 60
MDRIREARRSTMAANGLTRRRHRTNSLRDSPDDDGGMEMQEPTRLRDRGSGKKERDRERE
Sbjct: 607 MDRIREARRSTMAANGLTRRRHRTNSLRDSPDDDGGMEMQEPTRLRDRGSGKKERDRERE 428
Query: 61 RERERDRLGRNKRRRNDRLMHGVREDGGEDTSEESINDEEDDDDEDGGGGGGGGSVRMLP 120
RERERDRLGRNKRRRNDRLMHGVREDGGEDTSEESINDEEDDDDEDGGGGGGGGSVRMLP
Sbjct: 427 RERERDRLGRNKRRRNDRLMHGVREDGGEDTSEESINDEEDDDDEDGGGGGGGGSVRMLP 248
Query: 121 LNPSTLSSSSSLTNHHHRKSFPPAKVFRPTPPTTWKAADEMIGVSVPRKARSASTKRSHE 180
LNPSTLSSSSSLTNHHHRKSFPPAKVFRPTPPTTWKAADEMIGVSVPRKARSASTKRSHE
Sbjct: 247 LNPSTLSSSSSLTNHHHRKSFPPAKVFRPTPPTTWKAADEMIGVSVPRKARSASTKRSHE 68
Query: 181 CWASSGGGIVPEQNHHRQPSSS 202
CWASSGGGIVPEQNHHRQPSSS
Sbjct: 67 CWASSGGGIVPEQNHHRQPSSS 2
Score = 31.2 bits (69), Expect = 3.1
Identities = 13/53 (24%), Positives = 25/53 (46%)
Frame = -1
Query: 936 GSSSSQKHAQNQQQKANNASGSNGGGSGSLQGFPVTKNPQSQILQLQQQQHRQ 988
G + + + N ++ ++ G GGG GS++ P+ + S L HR+
Sbjct: 349 GGEDTSEESINDEEDDDDEDGGGGGGGGSVRMLPLNPSTLSSSSSLTNHHHRK 191
>BE123942 similar to GP|14194103|gb AT3g22390/MCB17_12 {Arabidopsis thaliana},
partial (1%)
Length = 516
Score = 329 bits (843), Expect = 6e-90
Identities = 170/171 (99%), Positives = 170/171 (99%)
Frame = +2
Query: 1213 SQQQMQRNQQILQLQKQNQFAAAAVAAARNKTPSTSNGSIYSDNLPSTSSISTKFPNAVS 1272
SQQQMQRNQQILQL KQNQFAAAAVAAARNKTPSTSNGSIYSDNLPSTSSISTKFPNAVS
Sbjct: 2 SQQQMQRNQQILQLLKQNQFAAAAVAAARNKTPSTSNGSIYSDNLPSTSSISTKFPNAVS 181
Query: 1273 AFPQSLVQSSNTVVTQSSQWKNSARVTNTSLSPQTMASPPSSSVKNPPQQQARSQQGHTQ 1332
AFPQSLVQSSNTVVTQSSQWKNSARVTNTSLSPQTMASPPSSSVKNPPQQQARSQQGHTQ
Sbjct: 182 AFPQSLVQSSNTVVTQSSQWKNSARVTNTSLSPQTMASPPSSSVKNPPQQQARSQQGHTQ 361
Query: 1333 ISFAANPKSSTPQVQTASSTQSPSPPVMVGSPTNSSMSKNTGSPRTTNSTS 1383
ISFAANPKSSTPQVQTASSTQSPSPPVMVGSPTNSSMSKNTGSPRTTNSTS
Sbjct: 362 ISFAANPKSSTPQVQTASSTQSPSPPVMVGSPTNSSMSKNTGSPRTTNSTS 514
Score = 34.3 bits (77), Expect = 0.37
Identities = 42/151 (27%), Positives = 54/151 (34%)
Frame = +2
Query: 795 LNQQQAAAAAAAASVRPGSVKSLPVTSNGPPSSTTNSAPPNTSGAGAAAPAPPTMSFTYP 854
L Q Q AAAA AA+ P TSNG S + P+TS ++S +P
Sbjct: 44 LKQNQFAAAAVAAARN-----KTPSTSNG---SIYSDNLPSTS----------SISTKFP 169
Query: 855 NMSGNETQYMAILQNNAYPFPIPAHVGGPPGYRGNPAQAFPFFNGSFYPSQMIHPSQIQS 914
N Q + N V N A+ N S P M P
Sbjct: 170 NAVSAFPQSLVQSSNT---------VVTQSSQWKNSARVT---NTSLSPQTMASPPSSSV 313
Query: 915 QQPPAQSQQSQQGHPNTTISTGSSSSQKHAQ 945
+ PP Q +SQQGH + + SS Q
Sbjct: 314 KNPPQQQARSQQGHTQISFAANPKSSTPQVQ 406
>TC89548 similar to GP|7300093|gb|AAF55262.1| CG10278 gene product {Drosophila
melanogaster}, partial (4%)
Length = 922
Score = 155 bits (392), Expect(2) = 1e-40
Identities = 81/94 (86%), Positives = 82/94 (87%), Gaps = 12/94 (12%)
Frame = +2
Query: 1391 TSSLSSQQPKNSPTMPTRKSSPVGGRNVPSILSGPQITSSNTG------------KQQQH 1438
TSSLSSQQPKNSPTMPTRKSSPVGGRNVPSILSGPQITSSNTG +QQQH
Sbjct: 2 TSSLSSQQPKNSPTMPTRKSSPVGGRNVPSILSGPQITSSNTGSKSQLSQQQQKQQQQQH 181
Query: 1439 QQISKQNLQQAQMFFSNPYMHSQVTQSNSPTSTT 1472
QQISKQNLQQAQMFFSNPYMHSQVTQSNSPTSTT
Sbjct: 182 QQISKQNLQQAQMFFSNPYMHSQVTQSNSPTSTT 283
Score = 38.5 bits (88), Expect = 0.020
Identities = 38/161 (23%), Positives = 62/161 (37%), Gaps = 14/161 (8%)
Frame = +2
Query: 1260 TSSISTKFPNAVSAFPQSLVQSSNTVVTQSSQWKNSARVTNTSLSPQTMASPPSSSVKNP 1319
TSS+S++ P ++S T+ T+ S V + PQ +S S +
Sbjct: 2 TSSLSSQQP-----------KNSPTMPTRKSSPVGGRNVPSILSGPQITSSNTGSKSQLS 148
Query: 1320 PQQQARSQQGHTQIS---------FAANPKSSTPQVQTASSTQSPSPP-----VMVGSPT 1365
QQQ + QQ H QIS F +NP + Q+ S T + S + P
Sbjct: 149 QQQQKQQQQQHQQISKQNLQQAQMFFSNPYMHSQVTQSNSPTSTTSATTGPYYLQRRGPE 328
Query: 1366 NSSMSKNTGSPRTTNSTSTNNKTSQTSSLSSQQPKNSPTMP 1406
+ +G + + + N+K S ++ P MP
Sbjct: 329 QQMQRQGSGGTSSNGAAANNSKGSTLNTQGLLHPSQFGAMP 451
Score = 31.2 bits (69), Expect(2) = 1e-40
Identities = 12/12 (100%), Positives = 12/12 (100%)
Frame = +1
Query: 1474 SYNWTVLSSTAW 1485
SYNWTVLSSTAW
Sbjct: 286 SYNWTVLSSTAW 321
>BG644598 weakly similar to PIR|T48107|T481 hypothetical protein F16M2.30 -
Arabidopsis thaliana, partial (2%)
Length = 827
Score = 95.1 bits (235), Expect(2) = 4e-23
Identities = 68/208 (32%), Positives = 103/208 (48%), Gaps = 10/208 (4%)
Frame = +2
Query: 608 QTNLEKNVQSNSLPIPMNVPSWPGGLPSMGYMT-------PLQGVVSMDGTTMPSAAIPP 660
++ +EK S+S+P+P V P L +G +T PLQ VV M+ TT S +
Sbjct: 32 KSQVEKTACSSSVPLPAAVSGLPSSLSPIGSITMTGGCTPPLQTVVKMNTTTGSSTSAQR 211
Query: 661 PHLLFNQPRPKRCATHCHIAQKILYNQQIARMNPFWPAAAGSASLYGAKPGS---NLSVV 717
+ ++P+PKRCATH IA+KIL +Q +MNP AA GS SL K + LS
Sbjct: 212 VNFALSEPQPKRCATHYQIARKILL-KQFTKMNPLLSAAIGSGSLCAPKTNNVNCLLSAE 388
Query: 718 PVPSTELHSGNIHGRATNSTQDKGPSLAMFPGHIGKDKSSQPSNVDNSSRKPILLQQTLP 777
+ + ++ N TQ+K ++ S+ P+ VD+ + +L Q P
Sbjct: 389 SMIINKQSQKHLPSLDQNGTQEKRLTVIGDHNLTATKNSNNPNPVDSIHQMHLLQQG--P 562
Query: 778 SGAAPSNILHGPTFIFPLNQQQAAAAAA 805
+ N++HGP F+F QA+ AAA
Sbjct: 563 FAGSTGNLVHGPAFVFSPAHYQASVAAA 646
Score = 32.7 bits (73), Expect(2) = 4e-23
Identities = 18/59 (30%), Positives = 29/59 (48%)
Frame = +3
Query: 809 VRPGSVKSLPVTSNGPPSSTTNSAPPNTSGAGAAAPAPPTMSFTYPNMSGNETQYMAIL 867
+R G V S TS+ S ++ + P T+ A MSF+YP+ S N + Y ++
Sbjct: 651 IRQGGVSSPNNTSSYNRSQSSVAGSPCTTSTLPATAIANAMSFSYPHFSANNSPYATLV 827
>CB893096
Length = 843
Score = 79.7 bits (195), Expect = 8e-15
Identities = 87/300 (29%), Positives = 135/300 (45%), Gaps = 10/300 (3%)
Frame = +1
Query: 1025 NIYGQNFAMPMQTPNFALMTTAMSGA--GSNGNHSEKKQQQQHPGSKAGGETSPA--FAM 1080
N+ GQNF +P+Q NF+ A S + G++GN +K+QQ K G E P+ FA+
Sbjct: 40 NLQGQNFTIPVQPVNFSFKPCATSDSVGGNSGNFGDKQQQ----ALKGGVEVIPSQPFAV 207
Query: 1081 PFPSITGA---TALDLSSIAQNHSIMQSSHNFQLMATAQAASAQL--KKSYHAAEEGKHV 1135
F S G + L+ SS+ QN ++QS + A+++ + +K+Y E K
Sbjct: 208 SFGSFNGTNLPSNLNFSSMKQNPLVIQSLPDVARQGYQAASTSHIVQQKNYSTTIE-KRG 384
Query: 1136 VNSSNLEEDRKAISGKIPTTGGQNIAFARSDVADPSMTSIACNNVIDSSGRSLNLGSASS 1195
NSS+ ++++K GK G + F S S N V+ S + S +S
Sbjct: 385 GNSSHKDDEKKTTHGKSSANGPTTLVFDNS--------SKNINFVLSHSNGNWPSHSITS 540
Query: 1196 RASASGMPSAINSNAAGSQQQMQRNQQILQLQKQNQFAAAAVAAA-RNKTPSTSNGSIYS 1254
+ + MP +SN + SQQ + Q LQL KQ+ AAA RNK ST+ S
Sbjct: 541 TVTTT-MP--FSSNTSSSQQ----SAQSLQLHKQHGMQQQQPAAATRNKASSTNTTS--- 690
Query: 1255 DNLPSTSSISTKFPNAVSAFPQSLVQSSNTVVTQSSQWKNSARVTNTSLSPQTMASPPSS 1314
+T F N F QS Q +++ T S+ + +P T P++
Sbjct: 691 ---------ATNFANNGPVFSQSHTQCNSSNQTSHSKITGRNHSYHVHHTPSTTIKTPTT 843
>BG451004 similar to GP|14194103|gb AT3g22390/MCB17_12 {Arabidopsis
thaliana}, partial (2%)
Length = 458
Score = 50.1 bits (118), Expect = 7e-06
Identities = 36/111 (32%), Positives = 55/111 (49%), Gaps = 3/111 (2%)
Frame = +3
Query: 666 NQPRPKRCATHCHIAQKILYNQQIARMNPFWPAAAGSASLYGAKPGS---NLSVVPVPST 722
++P+PKRCATH IA+KIL +Q +MNP AA GS SL K + LS +
Sbjct: 30 SEPQPKRCATHYQIARKILL-KQFTKMNPLLSAAIGSGSLCAPKTNNVNCLLSAESMIIN 206
Query: 723 ELHSGNIHGRATNSTQDKGPSLAMFPGHIGKDKSSQPSNVDNSSRKPILLQ 773
+ ++ N TQ+K ++ S+ P+ VD+ + +L Q
Sbjct: 207 KQSQKHLPSLDQNGTQEKRLTVIGDHNLTATKNSNNPNPVDSIHQMHLLQQ 359
>TC84279 weakly similar to GP|1799947|dbj|BAA16433.1 BENZENE 1 2-DIOXYGENASE
ALPHA SUBUNIT (EC 1.14.12.3). {Escherichia coli}, partial
(24%)
Length = 831
Score = 48.9 bits (115), Expect = 1e-05
Identities = 45/164 (27%), Positives = 74/164 (44%), Gaps = 1/164 (0%)
Frame = +3
Query: 1245 PSTSNGSIYSDNLPSTSSISTKFPNAVSAFPQSLVQSSNTVVTQSSQWKNSARVTNTSLS 1304
PS GS S P+TSS S++ A S +S+ + T S + T +S
Sbjct: 261 PSPGTGS--STTWPATSSSSSRTARAAST------RSTTSAGTGPSPSSRATAATASSSP 416
Query: 1305 PQTMASPPSSSVKNPPQQQARSQQGHTQISFAANPKSSTPQVQTASSTQSPSPPVMVGSP 1364
T A P +S+ P R+ + T A+ P +ST TASS + +PP SP
Sbjct: 417 ASTTAGPTASAATWPRPPGTRTSRASTSPRTASFPSTST-STPTASSGSTWTPPRRPRSP 593
Query: 1365 TNSSMSKNTG-SPRTTNSTSTNNKTSQTSSLSSQQPKNSPTMPT 1407
++++ +T S +T + +T T++ + ++SPT T
Sbjct: 594 GTTTLTASTSRSASSTTTLTTTTLTTRGRWRADTTGRSSPTTTT 725
Score = 33.5 bits (75), Expect = 0.63
Identities = 48/215 (22%), Positives = 78/215 (35%), Gaps = 4/215 (1%)
Frame = +3
Query: 122 NPSTLSSSSSLTNHHHRKSFPPAKVFRPTP----PTTWKAADEMIGVSVPRKARSASTKR 177
+P +SSS + S P + P+P TTW A S R AR+AST+
Sbjct: 180 HPRRCTSSSGAPSSPGNGS*RPTRPGCPSPGTGSSTTWPATSS----SSSRTARAASTRS 347
Query: 178 SHECWASSGGGIVPEQNHHRQPSSSPVRASAAPPSPSSSNASIRKKIKANGGGGGGGPKF 237
+ S+G G P +SS ++ A P+ S++ + +
Sbjct: 348 T----TSAGTGPSPSSRATAATASSSPASTTAGPTASAATWPRPPGTRTSRASTSPRTAS 515
Query: 238 RPPKSSSVTTKASSSVQDEIEIEIAEVLYGMMRQPQSQVAPSKQEMNDSIKMDSREINNN 297
P S+S T +S S + R+P+S P + S +
Sbjct: 516 FPSTSTSTPTASSGST------------WTPPRRPRS---PGTTTLTASTSRSASSTTTL 650
Query: 298 KSSASDSKSRISSPPQNSSSSATPVSAVVAPKRKR 332
++ ++ R + SS T SA A +R R
Sbjct: 651 TTTTLTTRGRWRADTTGRSSPTTTTSATTANRRTR 755
Score = 31.6 bits (70), Expect = 2.4
Identities = 27/106 (25%), Positives = 40/106 (37%)
Frame = +3
Query: 756 SSQPSNVDNSSRKPILLQQTLPSGAAPSNILHGPTFIFPLNQQQAAAAAAAASVRPGSVK 815
S PS+ N S +P T P +P G + +P ++ A AAS R
Sbjct: 204 SGAPSSPGNGS*RP-----TRPGCPSPGT---GSSTTWPATSSSSSRTARAASTR----- 344
Query: 816 SLPVTSNGPPSSTTNSAPPNTSGAGAAAPAPPTMSFTYPNMSGNET 861
S GP S+ +A +S + P + T+P G T
Sbjct: 345 STTSAGTGPSPSSRATAATASSSPASTTAGPTASAATWPRPPGTRT 482
>TC93234 weakly similar to GP|21751020|dbj|BAC03887. unnamed protein product
{Homo sapiens}, partial (11%)
Length = 591
Score = 46.6 bits (109), Expect = 7e-05
Identities = 50/178 (28%), Positives = 74/178 (41%), Gaps = 4/178 (2%)
Frame = +2
Query: 1241 RNKTPSTSNGSIYSDNLPSTSSISTKFPNAVSAFPQSLVQSSNTVVTQSSQWKNSA---- 1296
+N P + S S S +S S++ SA + SS+++ +Q S+ S
Sbjct: 71 KNCHPKSPKSSRNSQQKWSRTS*SSERATPASAS----LTSSSSIPSQRSRTSRSPSSRP 238
Query: 1297 RVTNTSLSPQTMASPPSSSVKNPPQQQARSQQGHTQISFAANPKSSTPQVQTASSTQSPS 1356
R T+T +P S P+SS P RS G T STP+ + S + P
Sbjct: 239 RHTSTGTAPPFAVSSPASSPTTP--SSTRSSPGST----------STPRTPSTSFSARPP 382
Query: 1357 PPVMVGSPTNSSMSKNTGSPRTTNSTSTNNKTSQTSSLSSQQPKNSPTMPTRKSSPVG 1414
P +P S +K + T +S+S TS S S P+ PTR+ SP G
Sbjct: 383 PSTPPPTPFRSRPTKGSSPSSTPSSSSPPAPTSPAISPS------RPSAPTRRPSPPG 538
Score = 30.8 bits (68), Expect = 4.1
Identities = 24/106 (22%), Positives = 40/106 (37%), Gaps = 11/106 (10%)
Frame = +2
Query: 753 KDKSSQPSNVDNSSRKPILLQQTLPSGAAPSNILHGP-TFIFPLNQQQAAAAAAAASVRP 811
+ SS+P + + P + S PS+ P + P + +A S P
Sbjct: 218 RSPSSRPRHTSTGTAPPFAVSSPASSPTTPSSTRSSPGSTSTPRTPSTSFSARPPPSTPP 397
Query: 812 ----------GSVKSLPVTSNGPPSSTTNSAPPNTSGAGAAAPAPP 847
GS S +S+ PP+ T+ + P+ A P+PP
Sbjct: 398 PTPFRSRPTKGSSPSSTPSSSSPPAPTSPAISPSRPSAPTRRPSPP 535
>TC81100 similar to PIR|H84824|H84824 En/Spm-like transposon protein
[imported] - Arabidopsis thaliana, partial (20%)
Length = 1266
Score = 46.6 bits (109), Expect = 7e-05
Identities = 41/152 (26%), Positives = 65/152 (41%), Gaps = 19/152 (12%)
Frame = +3
Query: 1301 TSLSPQTMASPPSSSVKNPPQQQARSQQGHTQISFAANPKSSTPQVQTA----SSTQSPS 1356
+S T S SS + P +RS + + + KSS P T+ +ST+S +
Sbjct: 681 SSFGSSTNGSKRISSSRGPSPATSRSSTPSGRPTLPSTTKSSRPSTPTSRATLTSTKSTA 860
Query: 1357 PPVMVGSPTNSSMSKNTGSPRTTNSTSTNNKTSQTSSLSSQQPKNS------PTMP--TR 1408
PPV +PT + +T + R + + ++ S T ++ S P + PT P T
Sbjct: 861 PPVRSSTPTRPTSRASTPTSRPSLTAPKTSQRSATPNIRSSTPSRTFGVSAPPTRPSSTS 1040
Query: 1409 KSSPVGGRN-------VPSILSGPQITSSNTG 1433
K+ PV +N PS+ S P S G
Sbjct: 1041 KARPVVAKNPVQSRGISPSVKSRPWEPSQMPG 1136
Score = 30.8 bits (68), Expect = 4.1
Identities = 50/241 (20%), Positives = 85/241 (34%), Gaps = 36/241 (14%)
Frame = +3
Query: 193 QNHHRQPSSSPVRASAAPPSPSSSNASIRK---KIKANGGGGGGGPKFRPPKSSSVTTKA 249
+ H +P++ +R PSS + + K + + G G + + S T
Sbjct: 576 ETHSSRPTALKLRVDNIQIEPSSRSNVVSKHHASMSSFGSSTNGSKRISSSRGPSPATSR 755
Query: 250 SSSVQDEIEIEIAEVLYGMMRQPQSQVAPSKQEMNDSIKMDSREINNNKSSASDSKSRIS 309
SS+ PS + S SR + S A+ + ++ +
Sbjct: 756 SST-------------------------PSGRPTLPSTTKSSRP-STPTSRATLTSTKST 857
Query: 310 SPPQNSSSSATPVS---------AVVAPK--RKRPRPVKHEDENPAIFGVRSSPV--SSI 356
+PP SS+ P S ++ APK ++ P FGV + P SS
Sbjct: 858 APPVRSSTPTRPTSRASTPTSRPSLTAPKTSQRSATPNIRSSTPSRTFGVSAPPTRPSST 1037
Query: 357 SKAESD--------------------HPSKMEACSSNSDKNNQGSVPEIPANLAPVQPSP 396
SKA PS+M + ++ N + S+PE PA++ +P
Sbjct: 1038SKARPVVAKNPVQSRGISPSVKSRPWEPSQMPGFTHDAPSNLKKSLPERPASVTRNRPGV 1217
Query: 397 P 397
P
Sbjct: 1218P 1220
>AJ502078 similar to GP|15529165|gb AT3g04880/T9J14_17 {Arabidopsis thaliana},
partial (53%)
Length = 647
Score = 46.2 bits (108), Expect = 9e-05
Identities = 38/115 (33%), Positives = 54/115 (46%), Gaps = 11/115 (9%)
Frame = +3
Query: 1292 WKNSARVTNTSLSPQTMA----------SPPSSSVKNPPQQQARSQQGHTQISFAANPKS 1341
W+N A + +SL+P T A S PS+S + + ++ + NP
Sbjct: 15 WQNPANTSKSSLAPMTSAHLSKMHYSLTSVPSTSKSKISEPLLTTPPVPLSVAASLNPSP 194
Query: 1342 STPQVQTASSTQSPSPPVMVGSPTNS-SMSKNTGSPRTTNSTSTNNKTSQTSSLS 1395
P+ SS +P+ PV SPTNS + S SP T ST + T+QTSSLS
Sbjct: 195 LPPKKSAVSSPAAPA-PVFPSSPTNSPACSPPLVSPLLTPST-PDQSTTQTSSLS 353
>BQ750934 similar to PIR|T19673|T19 hypothetical protein C33B4.3 -
Caenorhabditis elegans, partial (1%)
Length = 627
Score = 45.4 bits (106), Expect = 2e-04
Identities = 37/150 (24%), Positives = 66/150 (43%), Gaps = 7/150 (4%)
Frame = +2
Query: 1257 LPSTSSISTKFPNAVSAFPQSLVQSSNTVVTQSSQWKNSARVTNTSLSPQTMASPPSSSV 1316
L ST++ S+ A + FP + ++ T+ ++ + V ++SLS T+++ SS
Sbjct: 164 LTSTTASSSVETTAAAVFPTTSAVANGTLTAVTASETVTDEVASSSLS--TLSATNSSVT 337
Query: 1317 KNPPQQQAR-------SQQGHTQISFAANPKSSTPQVQTASSTQSPSPPVMVGSPTNSSM 1369
P A S T + ++ P S+ + PP +V T S
Sbjct: 338 SAAPSFTAEAETTSPVSSASSTSLETLDATTTAVPSSSIPESSSTSLPPTLVPQNTTSPA 517
Query: 1370 SKNTGSPRTTNSTSTNNKTSQTSSLSSQQP 1399
S + +T S+S+++ +S TSS SS P
Sbjct: 518 SSPSSITTSTTSSSSSSSSSTTSSSSSSSP 607
Score = 33.9 bits (76), Expect = 0.48
Identities = 34/161 (21%), Positives = 72/161 (44%), Gaps = 5/161 (3%)
Frame = +2
Query: 1165 SDVADPSMTSIACNNVIDSSGRSLNLGSASSRASASGMPSAINSNAAGSQQQMQRNQQIL 1224
S D +TS ++ ++++ ++ ++ A A+G +A+ ++ + + + L
Sbjct: 143 SATEDSVLTSTTASSSVETTAAAV---FPTTSAVANGTLTAVTASETVTDEVASSSLSTL 313
Query: 1225 QLQKQNQFAAAA--VAAARNKTPSTSNGSIYSDNLPSTSSI--STKFPNAVS-AFPQSLV 1279
+ +AA A A +P +S S + L +T++ S+ P + S + P +LV
Sbjct: 314 SATNSSVTSAAPSFTAEAETTSPVSSASSTSLETLDATTTAVPSSSIPESSSTSLPPTLV 493
Query: 1280 QSSNTVVTQSSQWKNSARVTNTSLSPQTMASPPSSSVKNPP 1320
+ T S ++ +++S S + S SSS PP
Sbjct: 494 PQNTTSPASSPSSITTSTTSSSSSSSSSTTSSSSSSSPTPP 616
>TC77946 homologue to GP|10334491|emb|CAC10207. putative splicing factor
{Cicer arietinum}, complete
Length = 2061
Score = 43.9 bits (102), Expect = 5e-04
Identities = 34/109 (31%), Positives = 54/109 (49%), Gaps = 4/109 (3%)
Frame = +3
Query: 2 DRIREARRSTMAANGLTRRRHRTNSLRDSPDDDGGMEMQEPTRLR---DRGSGKKERDRE 58
D+ +++T + + R +R + + D D DGG E + R R D KK+RDR+
Sbjct: 150 DKDSSKKKNTDITDIKSERTYRKHGIDD--DLDGG-EDRRSKRSRGGDDENGSKKDRDRD 320
Query: 59 RERERERDRLGRNKRRRNDR-LMHGVREDGGEDTSEESINDEEDDDDED 106
R+RER+R+ R+ R +R G RE E E D+E D + +
Sbjct: 321 RDRERDREHRERSSGRHRERDGERGSREKEREKDVERGGRDKERDRERE 467
Score = 40.8 bits (94), Expect = 0.004
Identities = 22/59 (37%), Positives = 34/59 (57%)
Frame = +3
Query: 20 RRHRTNSLRDSPDDDGGMEMQEPTRLRDRGSGKKERDRERERERERDRLGRNKRRRNDR 78
R HR S + DG +E R +D G ++++R+RERE+ER+R R+K R +R
Sbjct: 339 REHRERSSGRHRERDGERGSREKEREKDVERGGRDKERDREREKERER--RDKEREKER 509
Score = 33.5 bits (75), Expect = 0.63
Identities = 25/77 (32%), Positives = 33/77 (42%), Gaps = 2/77 (2%)
Frame = +3
Query: 26 SLRDSPDDDGGMEMQEPTRLRDRGSGKKERDRERERERERD--RLGRNKRRRNDRLMHGV 83
S R DD ++ R RDR ++ R+R R RERD R R K R D
Sbjct: 261 SKRSRGGDDENGSKKDRDRDRDRERDREHRERSSGRHRERDGERGSREKEREKD------ 422
Query: 84 REDGGEDTSEESINDEE 100
E GG D + ++E
Sbjct: 423 VERGGRDKERDREREKE 473
Score = 32.3 bits (72), Expect = 1.4
Identities = 25/94 (26%), Positives = 41/94 (43%), Gaps = 13/94 (13%)
Frame = +3
Query: 23 RTNSLRDSPDDDGGMEMQEPTRLRDRGSGKKERD--RERERERER-----------DRLG 69
R+ R D++G + ++ R R+R +ER R RER+ ER +R G
Sbjct: 258 RSKRSRGGDDENGSKKDRDRDRDRERDREHRERSSGRHRERDGERGSREKEREKDVERGG 437
Query: 70 RNKRRRNDRLMHGVREDGGEDTSEESINDEEDDD 103
R+K R +R R D + E + E + +
Sbjct: 438 RDKERDREREKERERRDKEREKERERRDKEREKE 539
>TC80366 similar to GP|13877617|gb|AAK43886.1 protein kinase-like protein
{Arabidopsis thaliana}, partial (16%)
Length = 814
Score = 43.9 bits (102), Expect = 5e-04
Identities = 38/133 (28%), Positives = 53/133 (39%)
Frame = +3
Query: 1301 TSLSPQTMASPPSSSVKNPPQQQARSQQGHTQISFAANPKSSTPQVQTASSTQSPSPPVM 1360
+S P T +SPP S+ NPP S P T ++ SP P
Sbjct: 3 SSPPPATPSSPPPSTPANPPP-----------------VTPSAPPPSTPATPSSPPPSTT 131
Query: 1361 VGSPTNSSMSKNTGSPRTTNSTSTNNKTSQTSSLSSQQPKNSPTMPTRKSSPVGGRNVPS 1420
+P S+ S + SP TT + S + T S S+ P +S P+ SS P
Sbjct: 132 PSAPPPSTPSNSPPSPPTTPAISPPSGGGTTPSPPSRTPPSSDDSPSPPSS-----KTPP 296
Query: 1421 ILSGPQITSSNTG 1433
S P +S +TG
Sbjct: 297 PPSPPSSSSISTG 335
Score = 36.6 bits (83), Expect = 0.075
Identities = 19/46 (41%), Positives = 25/46 (54%)
Frame = +3
Query: 816 SLPVTSNGPPSSTTNSAPPNTSGAGAAAPAPPTMSFTYPNMSGNET 861
S P T + PP STT SAPP ++ + + P+PPT P G T
Sbjct: 90 STPATPSSPPPSTTPSAPPPSTPSN-SPPSPPTTPAISPPSGGGTT 224
Score = 34.7 bits (78), Expect = 0.28
Identities = 33/118 (27%), Positives = 49/118 (40%), Gaps = 2/118 (1%)
Frame = +3
Query: 822 NGPPSSTTNSAPPNTSGAGAAAPAPPTMSFTYPNMSGNETQYMAILQNNAYPFPIPAHVG 881
+ PP +T +S PP+T A P P T S P+ + +A P P++
Sbjct: 3 SSPPPATPSSPPPST----PANPPPVTPSAPPPSTPATPSSPPPSTTPSAPPPSTPSN-- 164
Query: 882 GPPGYRGNPAQAFPFFNGSF-YPSQMIHPSQIQSQQPPAQSQQSQQGHP-NTTISTGS 937
PP PA + P G+ P PS S PP+ P +++ISTG+
Sbjct: 165 SPPSPPTTPAISPPSGGGTTPSPPSRTPPSSDDSPSPPSSKTPPPPSPPSSSSISTGT 338
Score = 30.0 bits (66), Expect = 7.0
Identities = 22/74 (29%), Positives = 28/74 (37%)
Frame = +3
Query: 777 PSGAAPSNILHGPTFIFPLNQQQAAAAAAAASVRPGSVKSLPVTSNGPPSSTTNSAPPNT 836
PS PS P P N + A S G + S PPSS + +PP
Sbjct: 105 PSSPPPSTTPSAPPPSTPSNSPPSPPTTPAISPPSGGGTTPSPPSRTPPSSDDSPSPP-- 278
Query: 837 SGAGAAAPAPPTMS 850
S P+PP+ S
Sbjct: 279 SSKTPPPPSPPSSS 320
>TC88762 similar to GP|22136928|gb|AAM91808.1 unknown protein {Arabidopsis
thaliana}, partial (3%)
Length = 805
Score = 43.1 bits (100), Expect = 8e-04
Identities = 41/153 (26%), Positives = 61/153 (39%), Gaps = 10/153 (6%)
Frame = +3
Query: 1272 SAFPQSLVQ--SSNTVVTQSSQWKNSARVTNTSLSPQ------TMASPPSSSVKNPPQQQ 1323
S+ P + Q S N + Q+ N T+ S SP T SPP+S+ + P
Sbjct: 9 SSLPPGVAQTPSGNGIAPSPYQFNNP---TSPSSSPSASSPVSTTTSPPASTPASSP--- 170
Query: 1324 ARSQQGHTQISFAANPKSSTPQVQTASSTQSPSPPVMVGSP--TNSSMSKNTGSPRTTNS 1381
+S +P +STP +T SP P SP TNS + GS +
Sbjct: 171 ---------VSTTTSPPASTPASSPVPTTTSPPAPTPASSPVSTNSPTASPAGSLPAAAT 323
Query: 1382 TSTNNKTSQTSSLSSQQPKNSPTMPTRKSSPVG 1414
S ++ T+ + S SS P + T +G
Sbjct: 324 PSPSSTTAGSPSPSSTPPGTRSSNETTPGRSIG 422
>TC85286 similar to GP|3776572|gb|AAC64889.1| ESTs gb|R65052 gb|AA712146
gb|H76533 gb|H76282 gb|AA650771 gb|H76287
gb|AA650887 gb|N37383, partial (77%)
Length = 1947
Score = 43.1 bits (100), Expect = 8e-04
Identities = 27/77 (35%), Positives = 39/77 (50%)
Frame = -3
Query: 32 DDDGGMEMQEPTRLRDRGSGKKERDRERERERERDRLGRNKRRRNDRLMHGVREDGGEDT 91
D+ G + +++ R R G++ER+RERERERER+R +R R RE E
Sbjct: 895 DEGGRVGVRKGASCRIRHEGERERERERERERERERERERERERE-------RERERERE 737
Query: 92 SEESINDEEDDDDEDGG 108
E E + + E GG
Sbjct: 736 RERERERERERERERGG 686
Score = 37.0 bits (84), Expect = 0.057
Identities = 21/57 (36%), Positives = 29/57 (50%)
Frame = -2
Query: 31 PDDDGGMEMQEPTRLRDRGSGKKERDRERERERERDRLGRNKRRRNDRLMHGVREDG 87
P+ G E + ++ER+RERERERER+R R + R +R RE G
Sbjct: 854 PNSARGRERERERERERERERERERERERERERERER-EREREREREREREREREGG 687
Score = 33.5 bits (75), Expect = 0.63
Identities = 18/52 (34%), Positives = 25/52 (47%)
Frame = -1
Query: 18 TRRRHRTNSLRDSPDDDGGMEMQEPTRLRDRGSGKKERDRERERERERDRLG 69
TR R R + + E + ++ER+RERERERER+R G
Sbjct: 843 TRERERERERERERERERERERERERERERERERERERERERERERERERGG 688
Score = 32.0 bits (71), Expect = 1.8
Identities = 21/55 (38%), Positives = 30/55 (54%), Gaps = 1/55 (1%)
Frame = -2
Query: 13 AANGLTRRRHRTNSLRDSPDDDGGMEMQ-EPTRLRDRGSGKKERDRERERERERD 66
+A G R R R + + E + E R R+R ++ER+RERERERER+
Sbjct: 848 SARGRERERERERERERERERERERERERERERERER---ERERERERERERERE 693
Score = 31.2 bits (69), Expect = 3.1
Identities = 21/55 (38%), Positives = 26/55 (47%)
Frame = -2
Query: 57 RERERERERDRLGRNKRRRNDRLMHGVREDGGEDTSEESINDEEDDDDEDGGGGG 111
R RERERER+R R + R +R RE E E E + + E GG G
Sbjct: 842 RGRERERERER-EREREREREREREREREREREREREREREREREREREREGGPG 681
>TC89255 similar to GP|3135257|gb|AAC16457.1| unknown protein {Arabidopsis
thaliana}, partial (28%)
Length = 1420
Score = 42.0 bits (97), Expect = 0.002
Identities = 40/143 (27%), Positives = 56/143 (38%), Gaps = 11/143 (7%)
Frame = +3
Query: 1302 SLSPQTMASPPSSSVKN---PPQQQARSQ------QGHTQISFAANPKSSTPQVQTASST 1352
S+ PQ +S SSS+ N PP+QQ S TQIS PKS Q
Sbjct: 186 SILPQPKSSSSSSSLFNSLPPPKQQPSSDTPIDAPSNFTQISSLPKPKSQIHQQPPKRVV 365
Query: 1353 QSPSPPVMVGSPTNSSMSKNTGSPRTTNSTSTNNKTSQTSSLSSQ--QPKNSPTMPTRKS 1410
Q P + + PT + ++ +T S S P+NS T+ + S
Sbjct: 366 QFKPPIIPLPKPTQLEDEDDEEERNRRRKMESSIQTPSVKSFLSTIPAPRNSSTLGVQSS 545
Query: 1411 SPVGGRNVPSILSGPQITSSNTG 1433
S G R++ + TSS G
Sbjct: 546 SGSGRRSILETSTPAPETSSGGG 614
>BG448933 similar to PIR|T09648|T09 nucleolin homolog nuM1 - alfalfa, partial
(29%)
Length = 663
Score = 42.0 bits (97), Expect = 0.002
Identities = 43/158 (27%), Positives = 65/158 (40%), Gaps = 8/158 (5%)
Frame = +2
Query: 312 PQNSSSSATPVSA---VVAPKRKRPRPVKHEDENPAIFGVRSSPVSSISKAESDHPSKME 368
P++S SAT V A VV+P + + + +E + V + A K+
Sbjct: 83 PKSSKKSATKVDAAPVVVSPVKSGKKGKRQAEEEVKAVSAKKQKVEEV--AAKQKALKVV 256
Query: 369 ACSSNSDKNNQGSVPEIPANLAPVQPSPPEPVKPESNTSSDAKVLTEESEKQKDVGLSKE 428
+S + + S E P AP PS P K + + + +EES+ +E
Sbjct: 257 KKEESSSEESSESEDEQPVVKAPA-PSKKTPAKKGNVKKAQPETTSEESDSDSSSSDEEE 433
Query: 429 VVPPVS----PKKESSVLQAVDDVREDVK-ATKALKEK 461
V PVS K S+ + VD ED K A KA+ K
Sbjct: 434 VKKPVSKAVPSKNGSAPAKKVDTSDEDKKPAAKAVPSK 547
>TC77947 homologue to GP|10334491|emb|CAC10207. putative splicing factor
{Cicer arietinum}, partial (17%)
Length = 742
Score = 42.0 bits (97), Expect = 0.002
Identities = 28/84 (33%), Positives = 42/84 (49%), Gaps = 1/84 (1%)
Frame = +3
Query: 19 RRRHRTNSLRDSPDDDGGMEMQEPTRLRDRGSGKKERDRERER-ERERDRLGRNKRRRND 77
R R R+ + + D +E +E R R R + ERDRERER ER+R R R++ R+
Sbjct: 27 RERERSRRSKSRSERDREIEREERERSR-RSISRSERDREREREERDRSRRSRSRSERDF 203
Query: 78 RLMHGVREDGGEDTSEESINDEED 101
+ R ++T E + E D
Sbjct: 204 EMRDSRRFREKKETVEPEADPERD 275
Score = 36.6 bits (83), Expect = 0.075
Identities = 17/54 (31%), Positives = 29/54 (53%)
Frame = +3
Query: 53 KERDRERERERERDRLGRNKRRRNDRLMHGVREDGGEDTSEESINDEEDDDDED 106
+ER+RE+ERERER R +++ R+ + RE S + E + ++ D
Sbjct: 3 REREREKERERERSRRSKSRSERDREIEREERERSRRSISRSERDREREREERD 164
Score = 29.6 bits (65), Expect = 9.1
Identities = 17/53 (32%), Positives = 25/53 (47%)
Frame = +3
Query: 52 KKERDRERERERERDRLGRNKRRRNDRLMHGVREDGGEDTSEESINDEEDDDD 104
++ER++ERERER R R++R R R SE E ++ D
Sbjct: 6 EREREKERERERSRRSKSRSERDREIEREERERSRRSISRSERDREREREERD 164
>BQ151174 similar to GP|11036868|gb| PxORF73 peptide {Plutella xylostella
granulovirus}, partial (42%)
Length = 909
Score = 39.7 bits (91), Expect = 0.009
Identities = 49/221 (22%), Positives = 71/221 (31%), Gaps = 9/221 (4%)
Frame = +1
Query: 191 PEQNHHRQPSSSPVRASAAPPSPSSSNASIRKKIKANGGGGGGGPKFRPPK-----SSSV 245
P+ HH + +P PP P++ + R + GG G P RPP +
Sbjct: 280 PKPPHHEKRPRTPEPPGGRPPPPTTPKQA-RPTTQERGGARGARP--RPPPAPPQGTGGE 450
Query: 246 TTKASSSVQDEIEIEIAEVLYGMMRQPQSQVAP----SKQEMNDSIKMDSREINNNKSSA 301
TK Q E + E G R P + P K E + +
Sbjct: 451 DTKTRPGKQREPQQNPRERGTGHARTPGKKNTPPHTKKKGETHKKSPGGRTPPPTPRPGG 630
Query: 302 SDSKSRISSPPQNSSSSATPVSAVVAPKRKRPRPVKHEDENPAIFGVRSSPVSSISKAES 361
++ + PPQ K PR HE++ PA + P + K +
Sbjct: 631 GGARGAPAPPPQKKQRE--------GGKSGPPRAPGHEEKKPAATSRKRPPRGAREKNPA 786
Query: 362 DHPSKMEACSSNSDKNNQGSVPEIPANLAPVQPSPPEPVKP 402
H + E KN Q P P + PP P P
Sbjct: 787 AHAREREK-KKTPKKNPQNHNNNPPKTPPPQKKHPPPPKTP 906
Score = 32.7 bits (73), Expect = 1.1
Identities = 39/169 (23%), Positives = 56/169 (33%), Gaps = 44/169 (26%)
Frame = +3
Query: 120 PLNPSTLSSSSSLTNHHHRKSFPPAKVFRPTPPTTWKAA-------DEMIGVSVPRKARS 172
P N +T +++ + + PP PTPP T + D P A
Sbjct: 174 PPNQTTNTTNPKKQHTPTTPTHPPRPHTPPTPPPTTQTTPPREAPTDPGTTRGTPPTADD 353
Query: 173 ASTKRSHECWASSGGGIVP--------------EQNHHRQ--------------PSSSPV 204
A T +H G G P QN R+ P + P
Sbjct: 354 AQTSPAHHPGEGGGKGGAPPPPPRPPPRHGRGGHQNQTREAKRTTTEPQGKRDRPRTDP- 530
Query: 205 RASAAPPSPSSSNASIRKKIK---------ANGGGGGGGPKFRPPKSSS 244
R PP + +K+ + A GGGG GGP+ PPK ++
Sbjct: 531 RKKKHPPPHQKEGGNTQKEPRGEDPPPHTPAGGGGGPGGPRPPPPKKTA 677
Score = 29.6 bits (65), Expect = 9.1
Identities = 23/116 (19%), Positives = 36/116 (30%)
Frame = +1
Query: 1305 PQTMASPPSSSVKNPPQQQARSQQGHTQISFAANPKSSTPQVQTASSTQSPSPPVMVGSP 1364
P+T P +QAR + A P+ Q + + P P
Sbjct: 307 PRTPEPPGGRPPPPTTPKQARPTTQERGGARGARPRPPPAPPQGTGGEDTKTRPGKQREP 486
Query: 1365 TNSSMSKNTGSPRTTNSTSTNNKTSQTSSLSSQQPKNSPTMPTRKSSPVGGRNVPS 1420
+ + TG RT +T T + + P PT + G R P+
Sbjct: 487 QQNPRERGTGHARTPGKKNTPPHTKKKGETHKKSPGGRTPPPTPRPGGGGARGAPA 654
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.307 0.122 0.342
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 43,937,584
Number of Sequences: 36976
Number of extensions: 720019
Number of successful extensions: 15502
Number of sequences better than 10.0: 457
Number of HSP's better than 10.0 without gapping: 8004
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 12779
length of query: 1485
length of database: 9,014,727
effective HSP length: 108
effective length of query: 1377
effective length of database: 5,021,319
effective search space: 6914356263
effective search space used: 6914356263
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.6 bits)
S2: 65 (29.6 bits)
Medicago: description of AC137838.13


