

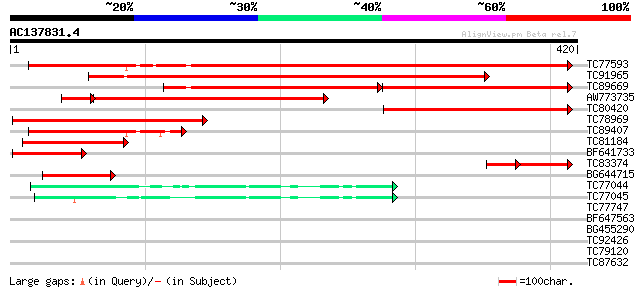
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137831.4 + phase: 0 /partial
(420 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC77593 homologue to GP|1237080|emb|CAA65539.1 ADP-glucose pyrop... 465 e-131
TC91965 similar to PIR|T06495|T06495 glucose-1-phosphate adenyly... 410 e-115
TC89669 homologue to GP|1237082|emb|CAA65540.1 ADP-glucose pyrop... 177 3e-89
AW773735 homologue to GP|13487785|gb ADP-glucose pyrophosphoryla... 243 3e-73
TC80420 homologue to GP|13487785|gb|AAK27719.1 ADP-glucose pyrop... 214 4e-56
TC78969 similar to GP|13487785|gb|AAK27719.1 ADP-glucose pyropho... 169 1e-42
TC89407 homologue to GP|1237082|emb|CAA65540.1 ADP-glucose pyrop... 139 3e-33
TC81184 similar to GP|13487783|gb|AAK27718.1 ADP-glucose pyropho... 122 3e-28
BF641733 weakly similar to GP|5852076|emb| ADP-glucose pyrophosp... 82 4e-16
TC83374 homologue to PIR|T08027|T08027 glucose-1-phosphate adeny... 57 4e-15
BG644715 similar to GP|4836911|gb|A 55420 {Arabidopsis thaliana}... 72 3e-13
TC77044 homologue to GP|4103324|gb|AAD01737.1| GDP-mannose pyrop... 47 1e-05
TC77045 homologue to PIR|T01007|T01007 mannose-1-phosphate guany... 43 3e-04
TC77747 similar to GP|3860325|emb|CAA10130.1 ribonuclease T2 {Ci... 32 0.46
BF647563 similar to GP|1237080|emb| ADP-glucose pyrophosphorylas... 31 0.78
BG455290 similar to GP|9049427|dbj| hypothetical protein {Oryza ... 30 1.7
TC92426 homologue to GP|10184858|emb|CAC09006. E. coli glgC16~Pe... 28 6.6
TC79120 similar to GP|14596089|gb|AAK68772.1 ATP-dependent Clp p... 28 6.6
TC87632 weakly similar to GP|21536611|gb|AAM60943.1 putative rib... 28 8.6
>TC77593 homologue to GP|1237080|emb|CAA65539.1 ADP-glucose
pyrophosphorylase {Pisum sativum}, partial (97%)
Length = 2119
Score = 465 bits (1196), Expect = e-131
Identities = 232/407 (57%), Positives = 306/407 (75%), Gaps = 4/407 (0%)
Frame = +3
Query: 15 KTVASIILGGGAGTRLFPLTQKRAKPAVPFGGCYRLVDIPMSNCINSEINKIYVLTQFNS 74
++V IILGGGAGTRL+PLT+KRAKPAVP G YRL+DIP+SNC+NS I+KIYVLTQFNS
Sbjct: 342 RSVLGIILGGGAGTRLYPLTKKRAKPAVPLGANYRLIDIPVSNCLNSNISKIYVLTQFNS 521
Query: 75 QSLNRHIARTY--NLGGGVNCGGSFIEVLAATQTLGESGNKWFQGTADAVRRFLWLFEDA 132
SLNRH++R Y N+GG N G F+EVLAA Q+ WFQGTADAVR++LWLFE+
Sbjct: 522 ASLNRHLSRAYASNMGGYKNEG--FVEVLAAQQS--PENPNWFQGTADAVRQYLWLFEE- 686
Query: 133 EHRNIENILVLCGDQLYRMDYMELVQKHINSCADISVSCLPVDGSRASDFGLVKVDERGR 192
N+ LVL GD LYRMDY +Q H S ADI+V+ LP+D +RA+ FGL+K+DE GR
Sbjct: 687 --HNVLEYLVLAGDHLYRMDYERFIQAHRESDADITVAALPMDEARATAFGLMKIDEEGR 860
Query: 193 IHQFMEKPKGDLLRSMHVDTSVFGLSAQEARKFPYIASMGIYVFKLDVLRKLLRSCYPNA 252
I +F EKPKG+ L++M VDT++ GL + A++ PYIASMGIYV V+ LLR +P A
Sbjct: 861 IIEFAEKPKGEQLKAMKVDTTILGLDDERAKEMPYIASMGIYVVSKHVMLDLLRDKFPGA 1040
Query: 253 NDFGSEVIPMAAK-DFKVQACLFNGYWEDIGTIKSFFDANLALMDKP-PKFQLYDQSKPI 310
NDFGSEVIP A + +VQA L++GYWEDIGTI++F++ANL + KP P F YD+S PI
Sbjct: 1041NDFGSEVIPGATELGMRVQAYLYDGYWEDIGTIEAFYNANLGITKKPIPDFSFYDRSSPI 1220
Query: 311 FTCPRFLPPTKLEKCQVVNSLVSDGCFLRECKVEHSIVGIRSRLNSGVQLKDTMMMGADY 370
+T PR+LPP+K+ + +S++ +GC ++ CK+ HS+VG+RS ++ G ++DT++MGADY
Sbjct: 1221YTQPRYLPPSKMLDADITDSVIGEGCVIKNCKIHHSVVGLRSCISEGAIIEDTLLMGADY 1400
Query: 371 YETEAEIASLLSAGDVPIGIGKNTKIMKCIIDKNARIGNNVTIANKE 417
YET+A+ L + G VPIGIGKN+ I + IIDKNARIG++V I N +
Sbjct: 1401YETDADRRFLAAKGSVPIGIGKNSHIKRAIIDKNARIGDDVKIINSD 1541
>TC91965 similar to PIR|T06495|T06495 glucose-1-phosphate
adenylyltransferase (EC 2.7.7.27) - garden pea, partial
(58%)
Length = 890
Score = 410 bits (1054), Expect = e-115
Identities = 198/297 (66%), Positives = 240/297 (80%)
Frame = +2
Query: 59 INSEINKIYVLTQFNSQSLNRHIARTYNLGGGVNCGGSFIEVLAATQTLGESGNKWFQGT 118
INS INKI+VLTQFNS SLNRHIARTY G G+N G ++EVLAATQT GE+G WFQGT
Sbjct: 2 INSGINKIFVLTQFNSASLNRHIARTY-FGNGINFGDGYVEVLAATQTPGEAGKNWFQGT 178
Query: 119 ADAVRRFLWLFEDAEHRNIENILVLCGDQLYRMDYMELVQKHINSCADISVSCLPVDGSR 178
ADAVR+F W+FEDA++ NIEN+++L GD LYRMDYM+LVQ HI+ ADI+VSC V SR
Sbjct: 179 ADAVRQFTWVFEDAKNTNIENVIILAGDHLYRMDYMDLVQSHIDRNADITVSCAAVGDSR 358
Query: 179 ASDFGLVKVDERGRIHQFMEKPKGDLLRSMHVDTSVFGLSAQEARKFPYIASMGIYVFKL 238
ASD+GLVKVD GRI QF EKPKG L+SM DTS+FGLS Q+A + PYIASMG+YVFK
Sbjct: 359 ASDYGLVKVDSGGRIIQFSEKPKGADLKSMQADTSLFGLSNQDALRSPYIASMGVYVFKT 538
Query: 239 DVLRKLLRSCYPNANDFGSEVIPMAAKDFKVQACLFNGYWEDIGTIKSFFDANLALMDKP 298
DVL KLL+ YP +NDFGSE+IP + K++ VQA F YWEDIGTIKSF+DAN+AL ++
Sbjct: 539 DVLLKLLKWRYPTSNDFGSEIIPASVKEYNVQAFFFGDYWEDIGTIKSFYDANMALTEES 718
Query: 299 PKFQLYDQSKPIFTCPRFLPPTKLEKCQVVNSLVSDGCFLRECKVEHSIVGIRSRLN 355
P F+ YD PIFT P FLPPTK++KC++V++++S GCFLREC V+HSIVG RSRL+
Sbjct: 719 PMFKFYDPKTPIFTSPGFLPPTKIDKCRIVDAIISHGCFLRECSVQHSIVGERSRLD 889
>TC89669 homologue to GP|1237082|emb|CAA65540.1 ADP-glucose
pyrophosphorylase {Pisum sativum}, partial (66%)
Length = 1438
Score = 177 bits (450), Expect(2) = 3e-89
Identities = 91/163 (55%), Positives = 119/163 (72%), Gaps = 1/163 (0%)
Frame = +1
Query: 115 FQGTADAVRRFLWLFEDAEHRNIENILVLCGDQLYRMDYMELVQKHINSCADISVSCLPV 174
FQGTADAVR++LWLFE+ N+ L+L GD LYRMDY + +Q H + ADI+V+ LP+
Sbjct: 319 FQGTADAVRQYLWLFEE---HNVLEYLILAGDHLYRMDYEKFIQAHRETDADITVAALPM 489
Query: 175 DGSRASDFGLVKVDERGRIHQFMEKPKGDLLRSMHVDTSVFGLSAQEARKFPYIASMGIY 234
D RA+ FGL+K+DE GRI +F EKPKGD L++M VDT++ GL + A++ P+IASMGIY
Sbjct: 490 DEKRATAFGLMKIDEEGRIIEFSEKPKGDQLQAMKVDTTILGLDDERAKEMPFIASMGIY 669
Query: 235 VFKLDVLRKLLRSCYPNANDFGSEVIPMAAKDFK-VQACLFNG 276
V +V+ LLR +P ANDFGSEVIP A K VQA L++G
Sbjct: 670 VISKNVMLDLLRDQFPGANDFGSEVIPGATSIGKRVQAYLYDG 798
Score = 169 bits (428), Expect(2) = 3e-89
Identities = 74/142 (52%), Positives = 108/142 (75%), Gaps = 1/142 (0%)
Frame = +2
Query: 277 YWEDIGTIKSFFDANLALMDKP-PKFQLYDQSKPIFTCPRFLPPTKLEKCQVVNSLVSDG 335
YWEDIGTI++F++ANL + KP P F YD+S PI+T PR+LPP+K+ + +S++ +G
Sbjct: 800 YWEDIGTIEAFYNANLGITKKPVPDFSFYDRSSPIYTQPRYLPPSKMLDADITDSVIGEG 979
Query: 336 CFLRECKVEHSIVGIRSRLNSGVQLKDTMMMGADYYETEAEIASLLSAGDVPIGIGKNTK 395
C ++ CK+ HS+VG+RS ++ G ++DT++MGADYYETEA+ + L + G VPIGIG+N+
Sbjct: 980 CVIKNCKIFHSVVGLRSCISEGAIIEDTLLMGADYYETEADKSFLAAKGSVPIGIGRNSH 1159
Query: 396 IMKCIIDKNARIGNNVTIANKE 417
I + I+DKNARIG NV I N +
Sbjct: 1160IKRAIVDKNARIGENVKIINSD 1225
>AW773735 homologue to GP|13487785|gb ADP-glucose pyrophosphorylase large
subunit CagpL2 {Cicer arietinum}, partial (37%)
Length = 602
Score = 243 bits (619), Expect(2) = 3e-73
Identities = 113/174 (64%), Positives = 146/174 (82%)
Frame = +1
Query: 63 INKIYVLTQFNSQSLNRHIARTYNLGGGVNCGGSFIEVLAATQTLGESGNKWFQGTADAV 122
+ +I++LTQ+NS SLNRH++R YN G G F+EVLAATQT GE+G KWFQGTADAV
Sbjct: 79 LEQIFILTQYNSFSLNRHLSRAYNFGNVSTFGEGFVEVLAATQTSGEAGKKWFQGTADAV 258
Query: 123 RRFLWLFEDAEHRNIENILVLCGDQLYRMDYMELVQKHINSCADISVSCLPVDGSRASDF 182
R+F+W+FEDA+++N+E+IL+L GD LYRM+YM+ VQKHI++ ADI+VSCLP+D SRASD+
Sbjct: 259 RQFIWVFEDAKNKNVEHILILSGDHLYRMNYMDFVQKHIDTNADITVSCLPMDDSRASDY 438
Query: 183 GLVKVDERGRIHQFMEKPKGDLLRSMHVDTSVFGLSAQEARKFPYIASMGIYVF 236
GL+K+DE GRI QF EKP G L++M VDT+V GLS +EA+K PYIASMG+YVF
Sbjct: 439 GLMKIDETGRIIQFAEKPXGSDLKAMRVDTTVLGLSPEEAKKNPYIASMGVYVF 600
Score = 50.8 bits (120), Expect(2) = 3e-73
Identities = 21/25 (84%), Positives = 23/25 (92%)
Frame = +3
Query: 39 KPAVPFGGCYRLVDIPMSNCINSEI 63
+PAVP GGCYRL+DIPMSNCINS I
Sbjct: 6 QPAVPIGGCYRLIDIPMSNCINSGI 80
>TC80420 homologue to GP|13487785|gb|AAK27719.1 ADP-glucose
pyrophosphorylase large subunit CagpL2 {Cicer
arietinum}, partial (33%)
Length = 777
Score = 214 bits (546), Expect = 4e-56
Identities = 97/140 (69%), Positives = 118/140 (84%)
Frame = +3
Query: 278 WEDIGTIKSFFDANLALMDKPPKFQLYDQSKPIFTCPRFLPPTKLEKCQVVNSLVSDGCF 337
WED+GTI+SFFD NLAL + PKFQ YD P FT PRFLPPTK+EKC++V++++S GCF
Sbjct: 15 WEDMGTIESFFDTNLALTEHAPKFQFYDPKTPFFTSPRFLPPTKVEKCKIVDAIISHGCF 194
Query: 338 LRECKVEHSIVGIRSRLNSGVQLKDTMMMGADYYETEAEIASLLSAGDVPIGIGKNTKIM 397
LREC V+HSIVGIRSRL SGV+L+DTMMMGADYY+TE+EIASLL+ G VP+G+G+NTKI
Sbjct: 195 LRECSVQHSIVGIRSRLESGVELQDTMMMGADYYQTESEIASLLAEGKVPVGVGENTKIR 374
Query: 398 KCIIDKNARIGNNVTIANKE 417
CIIDKNA+IG N I N +
Sbjct: 375 NCIIDKNAKIGRNAIITNAD 434
>TC78969 similar to GP|13487785|gb|AAK27719.1 ADP-glucose pyrophosphorylase
large subunit CagpL2 {Cicer arietinum}, partial (32%)
Length = 818
Score = 169 bits (429), Expect = 1e-42
Identities = 87/146 (59%), Positives = 108/146 (73%), Gaps = 2/146 (1%)
Frame = +3
Query: 3 QSPIFAGQEANPKTVASIILGGGAGTRLFPLTQKRAKPAVPFGGCYRLVDIPMSNCINSE 62
+ P F +A+PK+VASIILGGGAGTRLFPLT KRAKPAVP GGCYRL+DIPMSNCINS
Sbjct: 378 EGPYFETPKADPKSVASIILGGGAGTRLFPLTSKRAKPAVPIGGCYRLIDIPMSNCINSG 557
Query: 63 INKIYVLTQFNSQSLNRHIARTYNLGGGVNCGGSFIEVLAATQTLGESGNKWF-QGTADA 121
I KI++LTQ+NS SLNRH++R YN G G F+EVLAATQT GES K +
Sbjct: 558 IRKIFILTQYNSFSLNRHLSRAYNFGNVSTFGEGFVEVLAATQTSGESWEKMVPKEQLML 737
Query: 122 VRRFLWLF-EDAEHRNIENILVLCGD 146
V++F+W+F + ++ +IL+L GD
Sbjct: 738 VKQFIWVFLKMPXNKMFXHILILSGD 815
>TC89407 homologue to GP|1237082|emb|CAA65540.1 ADP-glucose
pyrophosphorylase {Pisum sativum}, partial (31%)
Length = 686
Score = 139 bits (349), Expect = 3e-33
Identities = 77/121 (63%), Positives = 94/121 (77%), Gaps = 4/121 (3%)
Frame = +1
Query: 15 KTVASIILGGGAGTRLFPLTQKRAKPAVPFGGCYRLVDIPMSNCINSEINKIYVLTQFNS 74
++V IILGGGAGTRL+PLT+KRAKPAVP G YRL+DIP+SNC+NS I+KIYVLTQFNS
Sbjct: 337 RSVLGIILGGGAGTRLYPLTKKRAKPAVPLGANYRLIDIPVSNCLNSNISKIYVLTQFNS 516
Query: 75 QSLNRHIARTY--NLGGGVNCGGSFIEVLAATQTLGES--GNKWFQGTADAVRRFLWLFE 130
SLNRH++R Y NLGG N G F+E LAA Q+ ES G++ ADAV ++LWLFE
Sbjct: 517 ASLNRHLSRAYASNLGGYKNEG--FVEDLAAQQSP*ESLIGSRVL---ADAVXQYLWLFE 681
Query: 131 D 131
+
Sbjct: 682 E 684
>TC81184 similar to GP|13487783|gb|AAK27718.1 ADP-glucose pyrophosphorylase
{Cicer arietinum}, partial (29%)
Length = 683
Score = 122 bits (305), Expect = 3e-28
Identities = 54/79 (68%), Positives = 66/79 (83%)
Frame = +1
Query: 10 QEANPKTVASIILGGGAGTRLFPLTQKRAKPAVPFGGCYRLVDIPMSNCINSEINKIYVL 69
++ +P TV ++ILGGGAGTRLFPLT++RAKPAVP GG YRL+D+PMSNCINS INK+Y+L
Sbjct: 424 EKRDPSTVVAVILGGGAGTRLFPLTKRRAKPAVPIGGAYRLIDVPMSNCINSGINKVYIL 603
Query: 70 TQFNSQSLNRHIARTYNLG 88
TQ NS +RH AR YN G
Sbjct: 604 TQVNSXLTHRHXARAYNFG 660
>BF641733 weakly similar to GP|5852076|emb| ADP-glucose pyrophosphorylase
{Ipomoea batatas}, partial (15%)
Length = 657
Score = 82.0 bits (201), Expect = 4e-16
Identities = 37/55 (67%), Positives = 45/55 (81%)
Frame = +3
Query: 3 QSPIFAGQEANPKTVASIILGGGAGTRLFPLTQKRAKPAVPFGGCYRLVDIPMSN 57
Q P F ++A+PK VASI+LGGG G +LFPLT++ A PAVP GGCYRL+DIPMSN
Sbjct: 492 QVPSFIRRKADPKNVASIVLGGGPGVQLFPLTKRAATPAVPVGGCYRLIDIPMSN 656
>TC83374 homologue to PIR|T08027|T08027 glucose-1-phosphate
adenylyltransferase (EC 2.7.7.27) large chain - Oriental
melon, partial (17%)
Length = 570
Score = 56.6 bits (135), Expect(2) = 4e-15
Identities = 26/42 (61%), Positives = 33/42 (77%)
Frame = +3
Query: 376 EIASLLSAGDVPIGIGKNTKIMKCIIDKNARIGNNVTIANKE 417
++ S L+ G VPIGIG N+K+ KCIIDKNARIG +V I NK+
Sbjct: 81 KLLSELAEGKVPIGIGSNSKVRKCIIDKNARIGKDVIIMNKD 206
Score = 42.4 bits (98), Expect(2) = 4e-15
Identities = 18/25 (72%), Positives = 24/25 (96%)
Frame = +2
Query: 354 LNSGVQLKDTMMMGADYYETEAEIA 378
L+ GV+L+DT+MMGADYY+TE+EIA
Sbjct: 14 LDYGVELQDTVMMGADYYQTESEIA 88
>BG644715 similar to GP|4836911|gb|A 55420 {Arabidopsis thaliana}, partial
(11%)
Length = 208
Score = 72.4 bits (176), Expect = 3e-13
Identities = 32/54 (59%), Positives = 43/54 (79%)
Frame = +3
Query: 25 GAGTRLFPLTQKRAKPAVPFGGCYRLVDIPMSNCINSEINKIYVLTQFNSQSLN 78
G+ ++L+PLT++R++ A+P G YRL+D +SNCINS I KIY LTQFNS SLN
Sbjct: 45 GSNSKLYPLTKRRSEAALPIAGNYRLIDAVISNCINSNIKKIYALTQFNSASLN 206
>TC77044 homologue to GP|4103324|gb|AAD01737.1| GDP-mannose
pyrophosphorylase {Solanum tuberosum}, complete
Length = 1631
Score = 47.0 bits (110), Expect = 1e-05
Identities = 65/273 (23%), Positives = 110/273 (39%), Gaps = 1/273 (0%)
Frame = +1
Query: 16 TVASIILGGGAGTRLFPLTQKRAKPAVPFGGCYRLVDIPMSNCINSEINKIYVLTQFNSQ 75
T+ ++IL GG GTRL PLT KP V F ++ + + +
Sbjct: 184 TMKALILVGGFGTRLRPLTLSVPKPLVDFANKPMILHQIEALKATGVTEVVLAINYQPEV 363
Query: 76 SLNRHIARTYNLGGGVNCGGSFIEVLAATQTLGESGNKWFQGTADAVRRFLWLFEDAEHR 135
LN LG ++C T+ LG +G A+ R L +D+
Sbjct: 364 MLNFLKDFEAKLGITISCSQE-------TEPLGTAGPL-------ALARDK-LIDDSG-- 492
Query: 136 NIENILVLCGDQLYRMDYMELVQKHINSCADISVSCLPVDGSRASDFGLVKVDER-GRIH 194
E VL D + E+++ H + + S+ VD S +G+V ++E G++
Sbjct: 493 --EPFFVLNSDVISDYPLKEMIEFHKSHGGEASIMVTKVD--EPSKYGVVVMEETTGQVE 660
Query: 195 QFMEKPKGDLLRSMHVDTSVFGLSAQEARKFPYIASMGIYVFKLDVLRKLLRSCYPNAND 254
+F+EKPK + V + + GIY+ VL ++ P + +
Sbjct: 661 KFVEKPK------LFVGNKI---------------NAGIYLLNPSVLDRI--ELRPTSIE 771
Query: 255 FGSEVIPMAAKDFKVQACLFNGYWEDIGTIKSF 287
E+ P A + K+ A + G+W DIG + +
Sbjct: 772 --KEIFPKIAAEKKLYAMVLPGFWMDIGQPRDY 864
>TC77045 homologue to PIR|T01007|T01007 mannose-1-phosphate
guanylyltransferase (EC 2.7.7.13) - Arabidopsis
thaliana, partial (80%)
Length = 997
Score = 42.7 bits (99), Expect = 3e-04
Identities = 64/277 (23%), Positives = 105/277 (37%), Gaps = 8/277 (2%)
Frame = +3
Query: 19 SIILGGGAGTRLFPLTQKRAKPAVPFGG-------CYRLVDIPMSNCINSEINKIYVLTQ 71
++IL GG GTRL PLT KP V F L DI ++ + + + V+
Sbjct: 132 ALILVGGFGTRLRPLTLSVPKPLVDFANKPMILHQIEALKDIGVTEVVLAINYQPEVMLN 311
Query: 72 FNSQSLNRHIARTYNLGGGVNCGGSFIEVLAATQTLGESGNKWFQGTADAVRRFLWLFED 131
F + L + C E L L + +K G+
Sbjct: 312 FLKDFEEK-------LDIKITCSQE-TEPLGTAGPLALARDKLIDGSG------------ 431
Query: 132 AEHRNIENILVLCGDQLYRMDYMELVQKHINSCADISVSCLPVDGSRASDFGLVKVDER- 190
E VL D + E+++ H + + S+ VD S +G+V ++E
Sbjct: 432 ------EPFFVLNSDVISEYPLKEMIKFHKSHGGEASIMVTKVD--EPSKYGVVVMEETT 587
Query: 191 GRIHQFMEKPKGDLLRSMHVDTSVFGLSAQEARKFPYIASMGIYVFKLDVLRKLLRSCYP 250
G++ +F+EKPK + V + + GIY+ VL + P
Sbjct: 588 GQVEKFVEKPK------LFVGNKI---------------NAGIYLLNPSVLDHI--ELRP 698
Query: 251 NANDFGSEVIPMAAKDFKVQACLFNGYWEDIGTIKSF 287
+ + EV P A + K+ A + G+W DIG + +
Sbjct: 699 TSIE--KEVFPKIAANKKLYAMVLPGFWMDIGQPRDY 803
>TC77747 similar to GP|3860325|emb|CAA10130.1 ribonuclease T2 {Cicer
arietinum}, partial (87%)
Length = 851
Score = 32.0 bits (71), Expect = 0.46
Identities = 16/48 (33%), Positives = 24/48 (49%), Gaps = 2/48 (4%)
Frame = +1
Query: 139 NILVLCGDQLYRMDYMELVQKHINSCADISVS--CLPVDGSRASDFGL 184
++ +LC Q + D+ VQ+ S D S C P G A+DFG+
Sbjct: 103 HVSILCASQSHDFDFFYFVQQWPGSYCDSQKSSCCYPTTGKPAADFGI 246
>BF647563 similar to GP|1237080|emb| ADP-glucose pyrophosphorylase {Pisum
sativum}, partial (10%)
Length = 654
Score = 31.2 bits (69), Expect = 0.78
Identities = 17/44 (38%), Positives = 27/44 (60%), Gaps = 1/44 (2%)
Frame = +1
Query: 262 MAAKDFKVQACLFNGYWEDIGTIKSFFDANLALMD-KPPKFQLY 304
+ K+ V A L++GY EDIG +++F +ANL + P F L+
Sbjct: 469 IGVKNSYV*AYLYDGYREDIGIVEAFNNANLGITK*HVPDFSLH 600
>BG455290 similar to GP|9049427|dbj| hypothetical protein {Oryza sativa
(japonica cultivar-group)}, partial (8%)
Length = 357
Score = 30.0 bits (66), Expect = 1.7
Identities = 20/67 (29%), Positives = 33/67 (48%), Gaps = 1/67 (1%)
Frame = -1
Query: 297 KPPKFQLYDQSKPIFTCPRFLPPTKLEKCQVVNSLVSD-GCFLRECKVEHSIVGIRSRLN 355
KPP+ Q+ +S F P+ P K K + S VSD GC+ K+ + +++
Sbjct: 321 KPPQIQITTESNLFFNIPKQTDPIKKHKKR*FFSNVSDLGCY----KLHFEFLRLKTYRG 154
Query: 356 SGVQLKD 362
GV+ K+
Sbjct: 153 EGVREKE 133
>TC92426 homologue to GP|10184858|emb|CAC09006. E. coli glgC16~Pea ssu TP
linked to E. coli glgC16 CDS {synthetic construct},
partial (12%)
Length = 159
Score = 28.1 bits (61), Expect = 6.6
Identities = 17/36 (47%), Positives = 22/36 (60%), Gaps = 3/36 (8%)
Frame = +3
Query: 228 IASMGIYVFKLDVLRKLLRSCYPNAN---DFGSEVI 260
+ASMGIYVF D L +LL + N DFG ++I
Sbjct: 51 LASMGIYVFDADYLYELL*EDDRDENSSHDFGKDLI 158
>TC79120 similar to GP|14596089|gb|AAK68772.1 ATP-dependent Clp
protease-like protein {Arabidopsis thaliana}, partial
(67%)
Length = 1110
Score = 28.1 bits (61), Expect = 6.6
Identities = 19/67 (28%), Positives = 32/67 (47%), Gaps = 3/67 (4%)
Frame = +1
Query: 16 TVASIILGGGAGTRLFPLTQKRAKPAVPFGGCYRL---VDIPMSNCINSEINKIYVLTQF 72
+ ASIILGGG + + R A P GG V+I ++++ N +++ F
Sbjct: 505 STASIILGGGTKGKRLAMPNTRIMIAQPPGGASGQAIDVEIQAKEIMHNKNNVSRIISSF 684
Query: 73 NSQSLNR 79
+SL +
Sbjct: 685 TGRSLEQ 705
>TC87632 weakly similar to GP|21536611|gb|AAM60943.1 putative ribosomal
protein S2 {Arabidopsis thaliana}, partial (81%)
Length = 1572
Score = 27.7 bits (60), Expect = 8.6
Identities = 18/54 (33%), Positives = 25/54 (45%), Gaps = 4/54 (7%)
Frame = +1
Query: 258 EVIPMAAKDFKVQACLFN--GYWEDIGTIKSFFDANLALMDKPP--KFQLYDQS 307
E++ KD K + L + W + TIK FD N + PP K YDQ+
Sbjct: 1387 EIVRNQDKDLKDKCKLIDTTNMWVSLRTIKRLFDTNRLKLQMPPISKEDDYDQT 1548
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.322 0.139 0.415
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,963,342
Number of Sequences: 36976
Number of extensions: 187204
Number of successful extensions: 794
Number of sequences better than 10.0: 38
Number of HSP's better than 10.0 without gapping: 742
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 786
length of query: 420
length of database: 9,014,727
effective HSP length: 99
effective length of query: 321
effective length of database: 5,354,103
effective search space: 1718667063
effective search space used: 1718667063
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 60 (27.7 bits)
Medicago: description of AC137831.4


