

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137831.14 + phase: 0
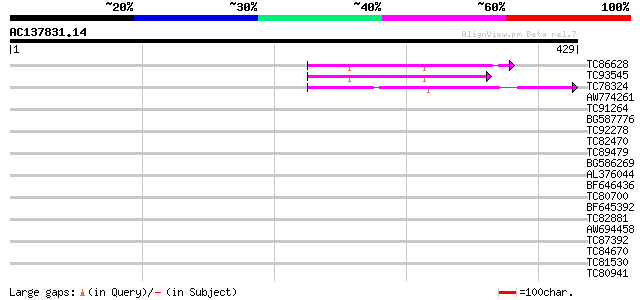
(429 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC86628 similar to PIR|T08971|T08971 hypothetical protein F19B15... 62 4e-10
TC93545 similar to PIR|F84581|F84581 copia-like retroelement pol... 55 4e-08
TC78324 similar to GP|8567798|gb|AAF76370.1| unknown protein {Ar... 52 6e-07
AW774261 weakly similar to EGAD|146423|156 vitellogenin {Anolis ... 35 0.056
TC91264 weakly similar to PIR|T27543|T27543 hypothetical protein... 33 0.16
BG587776 weakly similar to PIR|F86242|F86 unknown protein 98896... 33 0.16
TC92278 similar to SP|O43093|KINH_SYNRA Kinesin heavy chain (Syn... 33 0.21
TC82470 homologue to GP|23497460|gb|AAN37003.1 hypothetical prot... 32 0.36
TC89479 similar to GP|22136170|gb|AAM91163.1 unknown protein {Ar... 32 0.61
BG586269 weakly similar to PIR|T39903|T399 serine-rich protein -... 32 0.61
AL376044 similar to GP|18656155|db ryanodine receptor {Hemicentr... 30 1.4
BF646436 weakly similar to GP|9828634|gb F1N21.5 {Arabidopsis th... 30 1.8
TC80700 similar to GP|9759160|dbj|BAB09716.1 contains similarity... 30 1.8
BF645392 weakly similar to GP|7630034|em putative protein {Arabi... 30 2.3
TC82881 similar to GP|15216026|emb|CAC51423. amino acid permease... 30 2.3
AW694458 similar to GP|20161315|dbj hypothetical protein {Oryza ... 29 3.1
TC87392 similar to GP|9759312|dbj|BAB09818.1 CDK5 activator-bind... 29 4.0
TC84670 weakly similar to GP|18266212|gb|AAL67496.1 senescence-a... 29 4.0
TC81530 similar to GP|10177440|dbj|BAB10736. gb|AAC55944.1~gene_... 28 5.2
TC80941 weakly similar to PIR|T39903|T39903 serine-rich protein ... 28 6.8
>TC86628 similar to PIR|T08971|T08971 hypothetical protein F19B15.190 -
Arabidopsis thaliana, partial (88%)
Length = 743
Score = 62.0 bits (149), Expect = 4e-10
Identities = 40/167 (23%), Positives = 90/167 (52%), Gaps = 10/167 (5%)
Frame = +3
Query: 226 TIEKLQQQLDVIDRRCELSRKSAVASLHSG-------NRKLALRYARELKLVTQSREKCS 278
T++KL + L++++++ + K A A + N+K A++ + +L Q E+
Sbjct: 192 TLDKLNETLEMLEKKENVLLKKAAAEVEKAKEYTKRKNKKAAIQCLKRKRLYEQQVEQLG 371
Query: 279 SLLNRVEEVHGVVVDAESTKTVSEAMQIGARAIK--ENKISVEDVDICLRDLQESIDSQK 336
+ R+ + ++ A++T +A++ GA +K + +++DVD + ++ E ++ K
Sbjct: 372 NFQLRIHDQMIMLEGAKATTETVDALRTGAATMKAMQKATNIDDVDKTMDEINEQTENMK 551
Query: 337 EVEKALE-QTPSYTDINDEDIEEELEELELALEKEAQVDTLEKTTTS 382
++++AL + D +++++E ELEELE A E + L+ T T+
Sbjct: 552 QIQEALSAPIGAAADFDEDELEAELEELESA---ELEEQLLQPTFTA 683
>TC93545 similar to PIR|F84581|F84581 copia-like retroelement pol
polyprotein [imported] - Arabidopsis thaliana, partial
(72%)
Length = 661
Score = 55.5 bits (132), Expect = 4e-08
Identities = 34/149 (22%), Positives = 79/149 (52%), Gaps = 10/149 (6%)
Frame = +1
Query: 226 TIEKLQQQLDVIDRRCELSRKSAVASLHSG-------NRKLALRYARELKLVTQSREKCS 278
T+EKL + L +++++ + K A A + N++ A++ + +L Q E+
Sbjct: 202 TLEKLNETLGMLEKKESVLLKKAAAEVERAKEFTRAKNKRAAIQCLKRKRLYEQQVEQLG 381
Query: 279 SLLNRVEEVHGVVVDAESTKTVSEAMQIGARAIK--ENKISVEDVDICLRDLQESIDSQK 336
+ R+ + ++ A++T A++ GA A+K + + ++ DVD + ++ E ++ K
Sbjct: 382 NFQLRIHDQMIMLEGAKATTETVAALRTGAAAMKTMQQETNINDVDATMDEINEQTENMK 561
Query: 337 EVEKALE-QTPSYTDINDEDIEEELEELE 364
++++ L + D +++++E ELE LE
Sbjct: 562 QIQELLSAPIGAAADFDEDELEAELEXLE 648
>TC78324 similar to GP|8567798|gb|AAF76370.1| unknown protein {Arabidopsis
thaliana}, complete
Length = 1108
Score = 51.6 bits (122), Expect = 6e-07
Identities = 47/211 (22%), Positives = 94/211 (44%), Gaps = 7/211 (3%)
Frame = +1
Query: 226 TIEKLQQQLDV-IDRRCELSRKSAVASLHSGNRKLALRYARELKLVTQSREKCSSLLNRV 284
T+++ ++LDV ++R + +K+ + G + A+R ++ ++ R+ L N+
Sbjct: 256 TVDEKIKKLDVELNRYKDQIKKTRPGPMQDGIKARAMRVLKQKRMYEGQRDM---LYNQT 426
Query: 285 EEVHGVVVDAESTKTVSEAMQIGARAIKENK-----ISVEDVDICLRDLQESIDSQKEVE 339
+ V AE K + M A K+ K + ++D+D ++ + +D E++
Sbjct: 427 FNLDQVQFAAEGIKDAQQTMSALKSANKDLKGMMKTVKIQDIDNLQDEMMDLMDVSNEIQ 606
Query: 340 KALEQTPSY-TDINDEDIEEELEELELALEKEAQVDTLEKTTTSEEGNATLEASELLSDT 398
+ L ++ S DI++ED+ EL+ LE +E E+ EG + + SD
Sbjct: 607 ETLGRSYSVPDDIDEEDLMGELDALEADMENES------------EGVPSYLQPDKESDF 750
Query: 399 LSNLKLSDRPVGKSRTTHAASEGHKTANLAI 429
S L L P G++ H + L +
Sbjct: 751 DSELNLPSAPTGQTAVPHGRANAQTEDELGL 843
>AW774261 weakly similar to EGAD|146423|156 vitellogenin {Anolis pulchellus},
partial (40%)
Length = 495
Score = 35.0 bits (79), Expect = 0.056
Identities = 20/72 (27%), Positives = 38/72 (52%)
Frame = +1
Query: 313 ENKISVEDVDICLRDLQESIDSQKEVEKALEQTPSYTDINDEDIEEELEELELALEKEAQ 372
+ KI +E+ + +++E + E E+ +E+ + N+ED EEE EE E E+ +
Sbjct: 280 DEKIDLEEENDPEEEMEEIEYEEVEEEEEVEEIEEEVEENEEDAEEEEEE*EKEEEEVEE 459
Query: 373 VDTLEKTTTSEE 384
DT++ +E
Sbjct: 460 DDTMQNLDDDDE 495
>TC91264 weakly similar to PIR|T27543|T27543 hypothetical protein ZC395.10 -
Caenorhabditis elegans, partial (24%)
Length = 767
Score = 33.5 bits (75), Expect = 0.16
Identities = 21/67 (31%), Positives = 30/67 (44%)
Frame = +2
Query: 331 SIDSQKEVEKALEQTPSYTDINDEDIEEELEELELALEKEAQVDTLEKTTTSEEGNATLE 390
++D E LE T IND EE E E + +E ++ + EK SEE E
Sbjct: 398 ALDDSDEEMPGLETVDDTTKINDSTKEESSEISEESKSEEKEIKSEEKEIKSEEKEIKSE 577
Query: 391 ASELLSD 397
E+ S+
Sbjct: 578 EKEIKSE 598
>BG587776 weakly similar to PIR|F86242|F86 unknown protein 98896-95855
[imported] - Arabidopsis thaliana, partial (23%)
Length = 608
Score = 33.5 bits (75), Expect = 0.16
Identities = 38/145 (26%), Positives = 61/145 (41%), Gaps = 3/145 (2%)
Frame = +2
Query: 243 LSRKSAVASLHSGNRKLALRYARELKLVTQSREKCSSLLNRVEEVHGVVVDAESTKTVSE 302
+ +K+A+ K + ELKL+ + K RVEE VD ES +
Sbjct: 218 VEQKTAIEKQRKEEEKKRRQQEAELKLIAEETAK------RVEEAIRKRVD-ESLSSEEV 376
Query: 303 AMQIGARAIKENKISVEDVDICLRDLQESIDSQKEVEKAL---EQTPSYTDINDEDIEEE 359
++I R ++E + + D + + QKE E AL +Q ED+E
Sbjct: 377 RVEI-QRRLEEGRKKLND--------EVAAQLQKEKEAALIEAKQKEEQARKEKEDLERM 529
Query: 360 LEELELALEKEAQVDTLEKTTTSEE 384
LEE +E+ + + LE+ EE
Sbjct: 530 LEENRRKIEESQRREALEQQRREEE 604
>TC92278 similar to SP|O43093|KINH_SYNRA Kinesin heavy chain (Synkin).
{Syncephalastrum racemosum}, partial (10%)
Length = 709
Score = 33.1 bits (74), Expect = 0.21
Identities = 24/91 (26%), Positives = 47/91 (51%), Gaps = 3/91 (3%)
Frame = +3
Query: 316 ISVEDVDICLRDLQESIDSQKEVEKALEQTPSYTDINDEDIEEELEELEL---ALEKEAQ 372
+++E++ R+L ES K + EQT + +E + + +ELE+ LE E +
Sbjct: 126 LTMEELSTLRRELSES----KSLVTQHEQTINELHYENEHLTRKRDELEIRLTTLELEYE 293
Query: 373 VDTLEKTTTSEEGNATLEASELLSDTLSNLK 403
+ L+KT EE N ++ +E +++ L+
Sbjct: 294 -ELLDKTIAEEEANNNIDITETIAELRGKLE 383
>TC82470 homologue to GP|23497460|gb|AAN37003.1 hypothetical protein
{Plasmodium falciparum 3D7}, partial (8%)
Length = 1064
Score = 32.3 bits (72), Expect = 0.36
Identities = 34/136 (25%), Positives = 60/136 (44%), Gaps = 14/136 (10%)
Frame = +2
Query: 275 EKCSSLLNRVEEVHGVVVDAESTKT------------VSEAMQIGARA--IKENKISVED 320
E C L N+ +E+ V E+ K V++ Q+G I E+K ++
Sbjct: 221 ECCEDLENKKKEIRDVGRIIEARKKMQGKIDECVKDFVAKEGQLGLMEDLIGEHKKELKT 400
Query: 321 VDICLRDLQESIDSQKEVEKALEQTPSYTDINDEDIEEELEELELALEKEAQVDTLEKTT 380
++ LR + ++I QKE+E +++ + + E ++ELE KE Q++ K
Sbjct: 401 KELELRQVMDNISKQKELESQVKELVNDLVSKQKHFESHIKELE---SKERQLEGRLKEH 571
Query: 381 TSEEGNATLEASELLS 396
EE +EL S
Sbjct: 572 ELEEKEFEGRMNELES 619
>TC89479 similar to GP|22136170|gb|AAM91163.1 unknown protein {Arabidopsis
thaliana}, partial (16%)
Length = 1759
Score = 31.6 bits (70), Expect = 0.61
Identities = 44/168 (26%), Positives = 74/168 (43%), Gaps = 40/168 (23%)
Frame = +2
Query: 255 GNRKLALRYARELKLVTQSREKCSSLLNRVEEVHGV------------------VVDAES 296
GNR + A ++V ++ E+ + V++VH + +VDA+
Sbjct: 671 GNRNCEIVDADSEEIVEENIEEIVQIEKDVKKVHDIPFEESNTSFVLKGNSNSEIVDADR 850
Query: 297 TK----TVSEAMQIGARAIKENKISVEDVD--ICLRDLQ--ESID--SQKEVEKALEQTP 346
+ + E +QI K N ISVE+VD I L+ + E +D S+K VE +E
Sbjct: 851 EEIIEADIGEIVQIEKNVKKVNDISVEEVDTSIVLKGHKNCEIVDANSEKTVEANIEDMV 1030
Query: 347 SY-TDINDE-DIEEELEELELALEKEAQV----------DTLEKTTTS 382
D N E DI E + + L++++ V +TLE+ +S
Sbjct: 1031QIEKDANKEHDISIEYDNIRGVLKRKSNVYISSNVKQALETLERAISS 1174
>BG586269 weakly similar to PIR|T39903|T399 serine-rich protein - fission
yeast (Schizosaccharomyces pombe), partial (8%)
Length = 805
Score = 31.6 bits (70), Expect = 0.61
Identities = 32/111 (28%), Positives = 44/111 (38%), Gaps = 8/111 (7%)
Frame = +3
Query: 327 DLQESIDSQKEVEKALEQTPSYTDINDEDIEEELEELELALEKEAQVDTLEKTTT--SEE 384
D IDS + E S T I E +L+ A E E +++ E SE
Sbjct: 24 DAPRIIDSAHDDATMAEPDDSSTKNKPGWINGEHTDLKPAGETEDELEPAEVRVDELSEL 203
Query: 385 GNATLEASELLSDTLSNLKLSDR------PVGKSRTTHAASEGHKTANLAI 429
+ TLE EL TL +LSD P G++ A + HK N +
Sbjct: 204 SDTTLELYELSDTTLELSELSDTEDGASLPAGRNEPCSAQGKVHKKCNTGL 356
>AL376044 similar to GP|18656155|db ryanodine receptor {Hemicentrotus
pulcherrimus}, partial (0%)
Length = 525
Score = 30.4 bits (67), Expect = 1.4
Identities = 18/52 (34%), Positives = 26/52 (49%)
Frame = +2
Query: 327 DLQESIDSQKEVEKALEQTPSYTDINDEDIEEELEELELALEKEAQVDTLEK 378
+L D + + E LE + +DED EEE EE E +K AQ+ + K
Sbjct: 32 ELDSEGDLEGDEEMDLEDLEGLEEDDDEDGEEEEEESEQPAKKPAQIPNVGK 187
>BF646436 weakly similar to GP|9828634|gb F1N21.5 {Arabidopsis thaliana},
partial (11%)
Length = 659
Score = 30.0 bits (66), Expect = 1.8
Identities = 23/73 (31%), Positives = 41/73 (55%)
Frame = +1
Query: 325 LRDLQESIDSQKEVEKALEQTPSYTDINDEDIEEELEELELALEKEAQVDTLEKTTTSEE 384
L+ +E + Q+ E+ L++ T+ + ++ ELE L+LA E A LEK++ +E+
Sbjct: 193 LQQKEEILKLQQNEEERLKKEKQATE---DYLQRELETLQLAKESFAAEMELEKSSLAEK 363
Query: 385 GNATLEASELLSD 397
A E ++LL D
Sbjct: 364 --AQNEKNQLLLD 396
>TC80700 similar to GP|9759160|dbj|BAB09716.1 contains similarity to myosin
heavy chain~gene_id:MEE6.21 {Arabidopsis thaliana},
partial (4%)
Length = 958
Score = 30.0 bits (66), Expect = 1.8
Identities = 26/110 (23%), Positives = 52/110 (46%)
Frame = +2
Query: 297 TKTVSEAMQIGARAIKENKISVEDVDICLRDLQESIDSQKEVEKALEQTPSYTDINDEDI 356
TKT+ + + + +I N + D+ ++ L++ I S+ E ALE + S + ++
Sbjct: 152 TKTIPKNQK--SASIPHNSKEMTDLREKIKMLEDLIKSK---ETALEASASSYLEKEREL 316
Query: 357 EEELEELELALEKEAQVDTLEKTTTSEEGNATLEASELLSDTLSNLKLSD 406
+ ++EELE +E+ Q K + T + SE + D +L +
Sbjct: 317 QSKIEELEDKVEELNQSIASPKVVADKSFTTTSDGSEEVRDKSMETELKE 466
>BF645392 weakly similar to GP|7630034|em putative protein {Arabidopsis
thaliana}, partial (13%)
Length = 650
Score = 29.6 bits (65), Expect = 2.3
Identities = 31/113 (27%), Positives = 49/113 (42%), Gaps = 4/113 (3%)
Frame = +3
Query: 312 KENKISVED----VDICLRDLQESIDSQKEVEKALEQTPSYTDINDEDIEEELEELELAL 367
KE ISV+D +D + D E +D LE DINDEDI++ E +
Sbjct: 114 KEKGISVKDKKKLLDFNVLDPDEIVDP---ASLNLE-----LDINDEDIDDSASENSCSS 269
Query: 368 EKEAQVDTLEKTTTSEEGNATLEASELLSDTLSNLKLSDRPVGKSRTTHAASE 420
+ +L+ S++ + + LSD ++ L+ SD G R A +
Sbjct: 270 DD----SSLQPYDLSDDDSDLKKTISQLSDVVAALRKSDDADGVERALDVAEK 416
>TC82881 similar to GP|15216026|emb|CAC51423. amino acid permease AAP1
{Vicia faba var. minor}, partial (28%)
Length = 1132
Score = 29.6 bits (65), Expect = 2.3
Identities = 13/48 (27%), Positives = 28/48 (58%)
Frame = +2
Query: 314 NKISVEDVDICLRDLQESIDSQKEVEKALEQTPSYTDINDEDIEEELE 361
N +S + + + +DL+ +++ EVE ++E PS+ D D ++ L+
Sbjct: 605 NIVSNQTLPLTQKDLEHNLEFDMEVEHSIEAVPSHKDSKLYDDDDRLK 748
>AW694458 similar to GP|20161315|dbj hypothetical protein {Oryza sativa
(japonica cultivar-group)}, partial (14%)
Length = 655
Score = 29.3 bits (64), Expect = 3.1
Identities = 25/108 (23%), Positives = 42/108 (38%), Gaps = 11/108 (10%)
Frame = +2
Query: 326 RDLQESIDSQKEVEKALEQTPSYTDINDEDIE---------EELEELELALEKEAQVDTL 376
+ L S+ S+K+ + + Y D +D+D+E E+ + E A E++ DT
Sbjct: 179 KTLAPSMASKKKQSEGIALLSMYNDNSDDDMEDAEEVNRQGEQDDAAEHAAEEDLTADTY 358
Query: 377 EKTTTSEEGNATLE--ASELLSDTLSNLKLSDRPVGKSRTTHAASEGH 422
T T T E L + + + + KS T GH
Sbjct: 359 RMTVTDSGNEVTGEGFTPNRLFSPVQEQQRVELKISKSATLTIVDYGH 502
>TC87392 similar to GP|9759312|dbj|BAB09818.1 CDK5 activator-binding
protein-like {Arabidopsis thaliana}, partial (72%)
Length = 2056
Score = 28.9 bits (63), Expect = 4.0
Identities = 15/50 (30%), Positives = 31/50 (62%), Gaps = 1/50 (2%)
Frame = +2
Query: 203 ISLSAAALSGASNLDCDVLYLIWTIEKLQQQLDVIDRR-CELSRKSAVAS 251
+ L AAL N++ ++ Y ++K+QQQL +DR+ ++ R +A+++
Sbjct: 434 VFLGEAALIITQNVNYEIPYQRKQVQKIQQQLAELDRKEADIKRSAALSA 583
>TC84670 weakly similar to GP|18266212|gb|AAL67496.1 senescence-associated
putative protein {Narcissus pseudonarcissus}, partial
(36%)
Length = 961
Score = 28.9 bits (63), Expect = 4.0
Identities = 40/185 (21%), Positives = 83/185 (44%), Gaps = 8/185 (4%)
Frame = +3
Query: 244 SRKSAVASLHSGNRKLALRYARELKLVTQSREKCSSLLNRVEEVHGVVVDAESTKTVSEA 303
+R + +L N++LA + + +T ++++ L +V DAE + ++
Sbjct: 96 TRHREIQTLLHDNQRLATTHLALKQDLTATQQELRQLSAAAADVKAER-DAEVRRIYEKS 272
Query: 304 MQIGA--RAIKENKISVEDVDICLRDLQESIDSQKEVEKALEQTPSYTDINDED------ 355
+++ A RA+ K D+D D++E + +KE+ + L+ S + ED
Sbjct: 273 LKMDAEVRAVAAMK---SDLDQVRADVRELAEVRKELVEHLQSVQSELALAREDLKPLPI 443
Query: 356 IEEELEELELALEKEAQVDTLEKTTTSEEGNATLEASELLSDTLSNLKLSDRPVGKSRTT 415
I+ ++E L +++ EK T + LE + ++ +N+ + R V K R
Sbjct: 444 IKVDIEALRHEIQRGRSAIEFEKKTHAN----NLEHNRVMD---TNMIIMTREVEKLRAE 602
Query: 416 HAASE 420
A +E
Sbjct: 603 LANAE 617
>TC81530 similar to GP|10177440|dbj|BAB10736.
gb|AAC55944.1~gene_id:K9H21.2~similar to unknown protein
{Arabidopsis thaliana}, partial (24%)
Length = 1135
Score = 28.5 bits (62), Expect = 5.2
Identities = 38/149 (25%), Positives = 60/149 (39%), Gaps = 4/149 (2%)
Frame = +1
Query: 228 EKLQQQLDVIDRRC--ELSRKSAVA-SLHSGNRKLALRYARELKLVTQ-SREKCSSLLNR 283
EKL+ + + ++RR E +R A A + RK A E K + RE L
Sbjct: 430 EKLRIEREELERRQKEEKARLQAEAKAAEEARRKAEAEAAAEAKRKRELEREAARQALQN 609
Query: 284 VEEVHGVVVDAESTKTVSEAMQIGARAIKENKISVEDVDICLRDLQESIDSQKEVEKALE 343
+E+ V ES + + + + A + + K + D Q K LE
Sbjct: 610 MEKT---VEINESCQFLEDLEMLSA--VHDEKTPSFKEEASPDDHQNGFGGIKLQGNPLE 774
Query: 344 QTPSYTDINDEDIEEELEELELALEKEAQ 372
Q Y ++DED EEEL + K+ +
Sbjct: 775 QLGLYMKVDDEDEEEELPQSAAEPSKDVE 861
>TC80941 weakly similar to PIR|T39903|T39903 serine-rich protein - fission
yeast (Schizosaccharomyces pombe), partial (15%)
Length = 557
Score = 28.1 bits (61), Expect = 6.8
Identities = 29/97 (29%), Positives = 42/97 (42%), Gaps = 12/97 (12%)
Frame = +2
Query: 326 RDLQESIDSQKEVEKALEQTPSYTDI------------NDEDIEEELEELELALEKEAQV 373
++ +E I+++KE E ++E+ D ++E EEE EE E EKE
Sbjct: 107 KEEKEKIEAEKEAEGSIEEKEEGNDEEKTEVEEKEEGDDNEKSEEETEEKEEGDEKE--- 277
Query: 374 DTLEKTTTSEEGNATLEASELLSDTLSNLKLSDRPVG 410
E T EE EA E DT+ K D+ G
Sbjct: 278 -KTEAETEEEE-----EADE---DTIDKSKEEDKAEG 361
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.315 0.132 0.368
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,090,724
Number of Sequences: 36976
Number of extensions: 137385
Number of successful extensions: 736
Number of sequences better than 10.0: 45
Number of HSP's better than 10.0 without gapping: 727
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 732
length of query: 429
length of database: 9,014,727
effective HSP length: 99
effective length of query: 330
effective length of database: 5,354,103
effective search space: 1766853990
effective search space used: 1766853990
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 60 (27.7 bits)
Medicago: description of AC137831.14


