

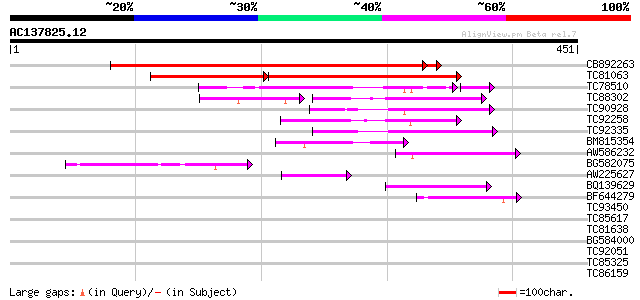
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137825.12 - phase: 0 /pseudo
(451 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
CB892263 similar to GP|13877621|gb Unknown protein {Arabidopsis ... 500 e-142
TC81063 similar to GP|13877621|gb|AAK43888.1 Unknown protein {Ar... 287 e-118
TC78510 similar to GP|6648201|gb|AAF21199.1| putative GTPase act... 56 3e-11
TC88302 similar to GP|8953714|dbj|BAA98077.1 contains similarity... 57 3e-11
TC90928 homologue to GP|21593435|gb|AAM65402.1 putative plant ad... 59 3e-09
TC92258 similar to GP|9758258|dbj|BAB08757.1 contains similarity... 55 6e-08
TC92335 similar to GP|14517422|gb|AAK62601.1 AT5g53570/MNC6_11 {... 54 9e-08
BM815354 similar to PIR|T05242|T05 hypothetical protein F18A5.12... 52 6e-07
AW586232 similar to PIR|T05242|T05 hypothetical protein F18A5.12... 50 2e-06
BG582075 similar to PIR|T05242|T052 hypothetical protein F18A5.1... 46 3e-05
AW225627 similar to GP|18377650|gb unknown protein {Arabidopsis ... 46 3e-05
BQ139629 similar to GP|15408841|dbj hypothetical protein~similar... 45 6e-05
BF644279 similar to PIR|T47641|T47 hypothetical protein T15C9.20... 41 8e-04
TC93450 similar to GP|8777354|dbj|BAA96944.1 microtubule-associa... 32 0.66
TC85617 similar to GP|2369766|emb|CAA04664.1 hypothetical protei... 32 0.66
TC81638 homologue to GP|10177686|dbj|BAB11012. emb|CAB62594.1~ge... 31 1.1
BG584000 weakly similar to GP|4097950|gb|A plant IF-like protein... 30 2.5
TC92051 similar to GP|4378659|gb|AAD19610.1| cyclic nucleotide g... 28 7.3
TC85325 28 7.3
TC86159 homologue to GP|21387177|gb|AAM47992.1 26S proteasome AA... 28 7.3
>CB892263 similar to GP|13877621|gb Unknown protein {Arabidopsis thaliana},
partial (47%)
Length = 804
Score = 500 bits (1287), Expect(2) = e-142
Identities = 248/252 (98%), Positives = 250/252 (98%)
Frame = +2
Query: 81 QISGAVEEEVQSTSKSVSAANVSKLKSSTSLGENLSEDIRKYTIGARATDSARVTKFNKV 140
QISGAVEEEVQSTSKSVSAANVSKLKSSTSLGENLSEDIRKYTIGARATDSARVTKFNKV
Sbjct: 2 QISGAVEEEVQSTSKSVSAANVSKLKSSTSLGENLSEDIRKYTIGARATDSARVTKFNKV 181
Query: 141 LSGTVVILDNLRELAWSGVPDYMRPKVWRLLLGYEPPNSDRKEGVLGRKRGEYLDCISQY 200
LSGTVVILDNLRELAWSGVPDYMRPKVWRLLLGYEPPNSDRKEGVLGRKRGEYLDCISQY
Sbjct: 182 LSGTVVILDNLRELAWSGVPDYMRPKVWRLLLGYEPPNSDRKEGVLGRKRGEYLDCISQY 361
Query: 201 YDIPDSERSDDEVNMLRQIAVDCPRTVPDVTFFQQQQVQKSLERILYAWAIRHPASGYVQ 260
YDIPDSERSDDEVNMLRQIAVDCPRTVPDVTFFQQQQVQKSLERILYAWAIRHPASGYVQ
Sbjct: 362 YDIPDSERSDDEVNMLRQIAVDCPRTVPDVTFFQQQQVQKSLERILYAWAIRHPASGYVQ 541
Query: 261 GINDLVTPFLVVFISEHLEGGIDDWSMSDLSSDKISNVEADCYWCLSKLLDGMQDHYTFA 320
GINDLVTPFLVVFISEHLEGGIDDWSMSDLSSDKISNVEADCYWCLSKLLDGMQ+HYTFA
Sbjct: 542 GINDLVTPFLVVFISEHLEGGIDDWSMSDLSSDKISNVEADCYWCLSKLLDGMQEHYTFA 721
Query: 321 QPGIQRLVFKLK 332
QPGIQRL F L+
Sbjct: 722 QPGIQRLGFNLR 757
Score = 25.0 bits (53), Expect(2) = e-142
Identities = 11/15 (73%), Positives = 13/15 (86%)
Frame = +1
Query: 329 FKLKELVRRIDEPIS 343
F +ELVR+IDEPIS
Sbjct: 745 F*FEELVRKIDEPIS 789
>TC81063 similar to GP|13877621|gb|AAK43888.1 Unknown protein {Arabidopsis
thaliana}, partial (53%)
Length = 749
Score = 287 bits (734), Expect(2) = e-118
Identities = 136/153 (88%), Positives = 147/153 (95%)
Frame = +2
Query: 207 ERSDDEVNMLRQIAVDCPRTVPDVTFFQQQQVQKSLERILYAWAIRHPASGYVQGINDLV 266
ERSDDE++MLRQIAVDCPRTVPDV FFQQ QVQKSLERILYAWAIRHPASGYVQGINDLV
Sbjct: 290 ERSDDEISMLRQIAVDCPRTVPDVAFFQQPQVQKSLERILYAWAIRHPASGYVQGINDLV 469
Query: 267 TPFLVVFISEHLEGGIDDWSMSDLSSDKISNVEADCYWCLSKLLDGMQDHYTFAQPGIQR 326
TPF VVF+SE+LEG I++W+MSDLSSDKISNVEADCYWCLSKLLDGMQDHYTFAQPGIQR
Sbjct: 470 TPFFVVFLSEYLEGSINNWTMSDLSSDKISNVEADCYWCLSKLLDGMQDHYTFAQPGIQR 649
Query: 327 LVFKLKELVRRIDEPISQHIEDQGLEFLQFAFR 359
LVFKLKELVRRID+P+S H+E+QGLEFL FAFR
Sbjct: 650 LVFKLKELVRRIDDPVSSHMENQGLEFLHFAFR 748
Score = 156 bits (394), Expect(2) = e-118
Identities = 75/94 (79%), Positives = 82/94 (86%)
Frame = +1
Query: 113 ENLSEDIRKYTIGARATDSARVTKFNKVLSGTVVILDNLRELAWSGVPDYMRPKVWRLLL 172
EN +E+IRK +IG R TDSARV KF KVLSGT+VILD LRELAWSGVPDYMRP VWRLLL
Sbjct: 7 ENPTEEIRKSSIGGRTTDSARVMKFTKVLSGTMVILDKLRELAWSGVPDYMRPTVWRLLL 186
Query: 173 GYEPPNSDRKEGVLGRKRGEYLDCISQYYDIPDS 206
GY P NSDR+EGVL RKR EYLDC+SQYYDIPD+
Sbjct: 187 GYAPTNSDRREGVLRRKRLEYLDCVSQYYDIPDT 288
>TC78510 similar to GP|6648201|gb|AAF21199.1| putative GTPase activator
protein {Arabidopsis thaliana}, partial (92%)
Length = 2077
Score = 55.8 bits (133), Expect(2) = 3e-11
Identities = 55/210 (26%), Positives = 86/210 (40%), Gaps = 4/210 (1%)
Frame = +2
Query: 151 LRELAWSGVPDYMRPKVWRLLLGYEPPNSDRKEGVLGRKRGEYLDCISQYYDIPDSERSD 210
L++L G+P +RPKVW L G +K+ D YYD
Sbjct: 917 LKKLIRKGIPPVLRPKVWFSLSG------------AAKKKSTVPD---SYYDDLTKAVEG 1051
Query: 211 DEVNMLRQIAVDCPRTVPDVTFFQQQQVQKSLERILYAWAIRHPASGYVQGINDLVTPFL 270
RQI D PRT P + + +L R+L A++ R GY QG+N + L
Sbjct: 1052 KVTPATRQIDHDLPRTFPGHAWLDTPEGHAALRRVLVAYSFRDSDVGYCQGLNYVAALLL 1231
Query: 271 VVFISEHLEGGIDDWSMSDLSSDKISNVEADCYWCLSKLLDG--MQDHYT--FAQPGIQR 326
+V E D +W L+ LL+ + D YT ++ +++
Sbjct: 1232 LVM-----------------------KTEEDAFWMLAVLLENVLVSDCYTNNLSRCHVEQ 1342
Query: 327 LVFKLKELVRRIDEPISQHIEDQGLEFLQF 356
VF K+L+ + I+ H++ LEF +F
Sbjct: 1343 RVF--KDLLTKKCPRIATHLD--SLEFDRF 1420
Score = 30.0 bits (66), Expect(2) = 3e-11
Identities = 10/27 (37%), Positives = 15/27 (55%)
Frame = +1
Query: 359 RWFNCLLIREIPFDLITRLWDTYLAEG 385
+WF CL + +P + R+WD EG
Sbjct: 1435 KWFLCLFSKSLPSETTLRVWDVIFYEG 1515
>TC88302 similar to GP|8953714|dbj|BAA98077.1 contains similarity to GTPase
activating protein~gene_id:F6N7.7 {Arabidopsis
thaliana}, complete
Length = 1198
Score = 57.0 bits (136), Expect(2) = 3e-11
Identities = 35/138 (25%), Positives = 61/138 (43%)
Frame = +1
Query: 242 LERILYAWAIRHPASGYVQGINDLVTPFLVVFISEHLEGGIDDWSMSDLSSDKISNVEAD 301
L IL ++ + GY QG++DL++P L V M D E++
Sbjct: 406 LRDILLTYSFYNFDLGYCQGMSDLLSPILFV--------------MED---------ESE 516
Query: 302 CYWCLSKLLDGMQDHYTFAQPGIQRLVFKLKELVRRIDEPISQHIEDQGLEFLQFAFRWF 361
+WC L++ + ++ Q G+ +F L +LV +D P+ + + + F FRW
Sbjct: 517 AFWCFVSLMERLGPNFNRDQNGMHSQLFALSKLVELLDSPLHNYFKQRDCLNYFFCFRWI 696
Query: 362 NCLLIREIPFDLITRLWD 379
RE ++ RLW+
Sbjct: 697 LIQFKREFEYEKTMRLWE 750
Score = 28.9 bits (63), Expect(2) = 3e-11
Identities = 25/87 (28%), Positives = 40/87 (45%), Gaps = 4/87 (4%)
Frame = +3
Query: 152 RELAWSGVPDYMRPKVWRLLLGYEPPNSD--RKEGVLGRKRGEYLDCISQYYDIPDSERS 209
RE + + R +VW LLLGY P +S +E + K+ EY +Q+ I ++
Sbjct: 117 REYFTADLTTNYRNEVWGLLLGYYPYDSTYAEREFLKSVKKSEYETIKNQWQSISSAQAK 296
Query: 210 DDEVNMLRQ--IAVDCPRTVPDVTFFQ 234
R+ I D RT +TF++
Sbjct: 297 RFTKFRERKGLIEKDVVRTDRSLTFYE 377
>TC90928 homologue to GP|21593435|gb|AAM65402.1 putative plant adhesion
molecule {Arabidopsis thaliana}, partial (66%)
Length = 1058
Score = 59.3 bits (142), Expect = 3e-09
Identities = 41/152 (26%), Positives = 67/152 (43%), Gaps = 5/152 (3%)
Frame = +2
Query: 239 QKSLERILYAWAIRHPASGYVQGINDLVTPFLVVFISEHLEGGIDDWSMSDLSSDKISNV 298
Q+SL +L A+++ GYVQG+ L L++++SE
Sbjct: 131 QRSLYNVLKAYSVFDREVGYVQGMGFLAG-LLLLYMSEE--------------------- 244
Query: 299 EADCYWCLSKLLDG-----MQDHYTFAQPGIQRLVFKLKELVRRIDEPISQHIEDQGLEF 353
D +W L LL G M+ Y P +Q+ +F+ + LVR + +H + +
Sbjct: 245 --DAFWLLVALLKGAVHAPMEGLYLAGLPLVQQYLFQFERLVREHLPKLGEHFTQEMINP 418
Query: 354 LQFAFRWFNCLLIREIPFDLITRLWDTYLAEG 385
+A +WF + PF L R+WD +L EG
Sbjct: 419 SMYASQWFITVFSYSFPFHLALRIWDVFLYEG 514
>TC92258 similar to GP|9758258|dbj|BAB08757.1 contains similarity to GTPase
activating protein~gene_id:MBG8.4 {Arabidopsis
thaliana}, partial (38%)
Length = 727
Score = 55.1 bits (131), Expect = 6e-08
Identities = 40/146 (27%), Positives = 68/146 (46%), Gaps = 2/146 (1%)
Frame = +1
Query: 216 LRQIAVDCPRTVPDVTFFQQQQVQKSLERILYAWAIRHPASGYVQGINDLVTPFLVVFIS 275
L QI +D RT + F+++Q+ L IL +A GY QG++DL +P +++
Sbjct: 349 LHQIGLDVVRTDRTLVFYEKQENLSKLWDILAVYAWIDKEVGYGQGMSDLCSPMIIL--- 519
Query: 276 EHLEGGIDDWSMSDLSSDKISNVEADCYWCLSKLLDGMQDHY--TFAQPGIQRLVFKLKE 333
+DD EAD +WC +L+ ++ ++ T G++ + L
Sbjct: 520 ------LDD--------------EADAFWCFERLMRRLRGNFRCTGRTLGVEAQLSNLAS 639
Query: 334 LVRRIDEPISQHIEDQGLEFLQFAFR 359
+ + ID + +HIE G FAFR
Sbjct: 640 ITQVIDPKLHKHIEHIGGGDYVFAFR 717
>TC92335 similar to GP|14517422|gb|AAK62601.1 AT5g53570/MNC6_11 {Arabidopsis
thaliana}, partial (37%)
Length = 1077
Score = 54.3 bits (129), Expect = 9e-08
Identities = 34/147 (23%), Positives = 65/147 (44%)
Frame = +1
Query: 242 LERILYAWAIRHPASGYVQGINDLVTPFLVVFISEHLEGGIDDWSMSDLSSDKISNVEAD 301
L IL A+A+ P GY QG++DL++P + V +H +
Sbjct: 13 LVAILEAYALYDPEIGYCQGMSDLLSPIICVVSEDH-----------------------E 123
Query: 302 CYWCLSKLLDGMQDHYTFAQPGIQRLVFKLKELVRRIDEPISQHIEDQGLEFLQFAFRWF 361
+WC + + ++ + GI+R + + ++++ D + +H+E E F +R
Sbjct: 124 AFWCFVGFMKKARQNFRLDEVGIRRQLDIVAKIIKFKDSHLFRHLEKLQAEDCFFVYRMV 303
Query: 362 NCLLIREIPFDLITRLWDTYLAEGDAL 388
L RE+ F+ LW+ A+ A+
Sbjct: 304 VVLFRRELTFEQTLCLWEVMWADQAAI 384
>BM815354 similar to PIR|T05242|T05 hypothetical protein F18A5.120 -
Arabidopsis thaliana, partial (31%)
Length = 494
Score = 51.6 bits (122), Expect = 6e-07
Identities = 32/111 (28%), Positives = 56/111 (49%), Gaps = 5/111 (4%)
Frame = +3
Query: 212 EVNMLRQIAVDCPRTVPDVTFF-----QQQQVQKSLERILYAWAIRHPASGYVQGINDLV 266
+ +++ QI D RT PD+ FF + Q++L+ IL +A +P YVQG+N+++
Sbjct: 177 DTDIIEQIDRDVKRTHPDMHFFCGDSQLAKSNQEALKNILIIFAKLNPGIRYVQGMNEVL 356
Query: 267 TPFLVVFISEHLEGGIDDWSMSDLSSDKISNVEADCYWCLSKLLDGMQDHY 317
P VF +D + + EAD ++C +LL G +D++
Sbjct: 357 APLFYVF-------------KNDPDEENAAFSEADTFFCFVELLSGFRDNF 470
>AW586232 similar to PIR|T05242|T05 hypothetical protein F18A5.120 -
Arabidopsis thaliana, partial (30%)
Length = 529
Score = 50.1 bits (118), Expect = 2e-06
Identities = 31/104 (29%), Positives = 54/104 (51%), Gaps = 5/104 (4%)
Frame = +1
Query: 308 KLLDGMQDHYTF----AQPGIQRLVFKLKELVRRIDEPISQHIEDQGLEFLQF-AFRWFN 362
+LL G++D++ + GI+ + KL +L+R+ DE + +H+E QF AFRW
Sbjct: 4 ELLSGLRDNFVQQLDNSVVGIRSTITKLSQLLRKHDEELWRHLEITSKINPQFYAFRWIT 183
Query: 363 CLLIREIPFDLITRLWDTYLAEGDALPDFLVYIFASFLLTTNSR 406
LL +E F +WDT L + + + L+ + + L+ R
Sbjct: 184 LLLTQEFNFADSLHIWDTLLGDPEGPQETLLRVCCAMLILIRKR 315
>BG582075 similar to PIR|T05242|T052 hypothetical protein F18A5.120 -
Arabidopsis thaliana, partial (21%)
Length = 784
Score = 46.2 bits (108), Expect = 3e-05
Identities = 43/151 (28%), Positives = 71/151 (46%), Gaps = 2/151 (1%)
Frame = +1
Query: 45 KVLMSRRSEPSDNLNSKVSSTFYKRSFSHNDAGTSDQISGAVEEEVQSTSKSVSAANVSK 104
K LM+++ P D LNS + S+ +++ + S+ S V+ +A S
Sbjct: 49 KPLMAKKRVP-DWLNSPMWSSPTEQNRFSATSYPSEPPSPPPVTVVEDPPSIHNAITTSP 225
Query: 105 LKSSTSLGENLSEDIRKYTIGARATDSARVTKFNKVLSGTVVILDNLRELAWSGVPDY-- 162
SS+S S D +I A +++ LS V+ + LR++A G+PD
Sbjct: 226 PSSSSSSASTSSTD--DMSISRLAQSQSQLLA---ELSRKVIDMRELRKIASQGIPDSPG 390
Query: 163 MRPKVWRLLLGYEPPNSDRKEGVLGRKRGEY 193
+R +W+LLLGY PP+ L +KR +Y
Sbjct: 391 LRSTIWKLLLGYLPPDRSLWSSELAKKRSQY 483
>AW225627 similar to GP|18377650|gb unknown protein {Arabidopsis thaliana},
partial (16%)
Length = 455
Score = 45.8 bits (107), Expect = 3e-05
Identities = 23/57 (40%), Positives = 33/57 (57%), Gaps = 1/57 (1%)
Frame = +3
Query: 217 RQIAVDCPRTVPDV-TFFQQQQVQKSLERILYAWAIRHPASGYVQGINDLVTPFLVV 272
R + D R P+ ++FQ Q L RIL W ++HP GY QG+++L+ PFL V
Sbjct: 42 RLVDQDLSRLYPEHGSYFQTPGCQGMLRRILLLWCLKHPDCGYRQGMHELLAPFLYV 212
>BQ139629 similar to GP|15408841|dbj hypothetical protein~similar to
Arabidopsis thaliana chromosome 2 At2g43490, partial
(15%)
Length = 666
Score = 45.1 bits (105), Expect = 6e-05
Identities = 24/85 (28%), Positives = 41/85 (48%), Gaps = 1/85 (1%)
Frame = +1
Query: 300 ADCYWCLSKLLDGMQDHYTFAQPG-IQRLVFKLKELVRRIDEPISQHIEDQGLEFLQFAF 358
AD +WC LL M++++ P + + + L ++ D+ + H+ G E L FAF
Sbjct: 34 ADAFWCFEMLLRRMRENFKMEGPTRVMKQLRALWHILELSDKELFAHLSKIGAESLHFAF 213
Query: 359 RWFNCLLIREIPFDLITRLWDTYLA 383
R L RE+ F+ +W+ A
Sbjct: 214 RMLLVLFRRELSFNEALSMWEVRAA 288
>BF644279 similar to PIR|T47641|T47 hypothetical protein T15C9.20 -
Arabidopsis thaliana, partial (20%)
Length = 681
Score = 41.2 bits (95), Expect = 8e-04
Identities = 24/89 (26%), Positives = 45/89 (49%), Gaps = 5/89 (5%)
Frame = +3
Query: 324 IQRLVFKLKELVRRIDEPISQHIEDQGLEFLQFAFRWFNCLLIREIPFDLITRLWDTYLA 383
+ +LVF+ ELVR ++ H+ G++ WF + + +P++ + R+WD L
Sbjct: 24 VDQLVFE--ELVRERFPKLANHLXYLGVQVAWVTGPWFLSIFVNMLPWESVLRVWDVLLF 197
Query: 384 EGDALPDF-----LVYIFASFLLTTNSRG 407
EG+ + F L+ ++ L+TT G
Sbjct: 198 EGNRVMLFXTAVALMELYGPALVTTKDAG 284
>TC93450 similar to GP|8777354|dbj|BAA96944.1 microtubule-associated
protein-like {Arabidopsis thaliana}, partial (22%)
Length = 806
Score = 31.6 bits (70), Expect = 0.66
Identities = 17/66 (25%), Positives = 26/66 (38%)
Frame = +3
Query: 322 PGIQRLVFKLKELVRRIDEPISQHIEDQGLEFLQFAFRWFNCLLIREIPFDLITRLWDTY 381
P + L L+ D + H+ D +E F RW L RE D + +WD
Sbjct: 84 PPVIEASMALYHLLSLADSSLHSHLLDLEVEPQYFYLRWLRVLFGREFSLDKLLVIWDEI 263
Query: 382 LAEGDA 387
A ++
Sbjct: 264 FASDNS 281
>TC85617 similar to GP|2369766|emb|CAA04664.1 hypothetical protein {Citrus x
paradisi}, partial (85%)
Length = 1489
Score = 31.6 bits (70), Expect = 0.66
Identities = 18/64 (28%), Positives = 30/64 (46%), Gaps = 2/64 (3%)
Frame = +1
Query: 1 MNKPPRENHERNNNNN--NTVTTLDSRFSQTLRNVQGLLKGRSMPGKVLMSRRSEPSDNL 58
+NK P N+ NNNNN N D RF +TL + L + + G + + ++L
Sbjct: 448 LNKDPHNNNSNNNNNNNENNSNATDKRF-KTLPAAETLPRNEVLGGYIFVCNNDTMQEDL 624
Query: 59 NSKV 62
++
Sbjct: 625 KRQL 636
>TC81638 homologue to GP|10177686|dbj|BAB11012.
emb|CAB62594.1~gene_id:MUL8.6~similar to unknown protein
{Arabidopsis thaliana}, partial (6%)
Length = 1585
Score = 30.8 bits (68), Expect = 1.1
Identities = 22/75 (29%), Positives = 39/75 (51%)
Frame = +2
Query: 34 QGLLKGRSMPGKVLMSRRSEPSDNLNSKVSSTFYKRSFSHNDAGTSDQISGAVEEEVQST 93
+ ++K + + +V ++R+ P + K S K SH TS + S + ++EV+S
Sbjct: 668 KSVVKEKHIVNEVKVNRKKTP---VVDKPPSLLSK---SHASPSTSQEFSSSTDKEVRSL 829
Query: 94 SKSVSAANVSKLKSS 108
SKS S + LKS+
Sbjct: 830 SKSTSTKVETPLKST 874
>BG584000 weakly similar to GP|4097950|gb|A plant IF-like protein
{Arabidopsis thaliana}, partial (24%)
Length = 833
Score = 29.6 bits (65), Expect = 2.5
Identities = 13/24 (54%), Positives = 16/24 (66%)
Frame = +1
Query: 8 NHERNNNNNNTVTTLDSRFSQTLR 31
NHE+NNNNNN V + + SQ R
Sbjct: 148 NHEKNNNNNNKVQYMKLKPSQQKR 219
>TC92051 similar to GP|4378659|gb|AAD19610.1| cyclic nucleotide gated
channel {Arabidopsis thaliana}, partial (25%)
Length = 665
Score = 28.1 bits (61), Expect = 7.3
Identities = 16/51 (31%), Positives = 22/51 (42%)
Frame = +3
Query: 353 FLQFAFRWFNCLLIREIPFDLITRLWDTYLAEGDALPDFLVYIFASFLLTT 403
F +F F W T+ D YL GD+L DF + + F+L T
Sbjct: 438 FAKFKFSW--------------TKPQDNYLCHGDSLCDFHCHFWIGFILIT 548
>TC85325
Length = 3584
Score = 28.1 bits (61), Expect = 7.3
Identities = 14/42 (33%), Positives = 25/42 (59%)
Frame = +3
Query: 360 WFNCLLIREIPFDLITRLWDTYLAEGDALPDFLVYIFASFLL 401
+ CL I E + IT++WD +L D + + +++F+ FLL
Sbjct: 2451 YLKCLEILESRYIKITKIWDLFL*HADWIVN--LFLFSIFLL 2570
>TC86159 homologue to GP|21387177|gb|AAM47992.1 26S proteasome AAA-ATPase
subunit RPT4a-like protein {Arabidopsis thaliana},
complete
Length = 1640
Score = 28.1 bits (61), Expect = 7.3
Identities = 28/110 (25%), Positives = 50/110 (45%), Gaps = 15/110 (13%)
Frame = +2
Query: 85 AVEEEVQSTSKSVSAANVSKLKSSTSLGENLSEDIR---------------KYTIGARAT 129
+V E ++++ K + LKS S+G+ + E +R +Y +G R+
Sbjct: 170 SVRENLRASKKEFNKTE-DDLKSLQSVGQIIGEVLRPLDNERLIVKASSGPRYVVGCRS- 343
Query: 130 DSARVTKFNKVLSGTVVILDNLRELAWSGVPDYMRPKVWRLLLGYEPPNS 179
+V K K+ SGT V+LD +P + P V+ +L +E P +
Sbjct: 344 ---KVDK-EKLTSGTRVVLDMTTLTIMRALPREVDPVVYNML--HEDPGN 475
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.320 0.135 0.411
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 14,107,710
Number of Sequences: 36976
Number of extensions: 195751
Number of successful extensions: 1293
Number of sequences better than 10.0: 42
Number of HSP's better than 10.0 without gapping: 1175
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1249
length of query: 451
length of database: 9,014,727
effective HSP length: 99
effective length of query: 352
effective length of database: 5,354,103
effective search space: 1884644256
effective search space used: 1884644256
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 60 (27.7 bits)
Medicago: description of AC137825.12


