

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137823.6 + phase: 0
(217 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
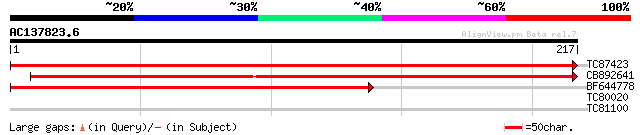
TC87423 weakly similar to GP|9758068|dbj|BAB08647.1 frnE protein... 439 e-124
CB892641 weakly similar to GP|9758068|dbj frnE protein-like {Ara... 390 e-109
BF644778 weakly similar to GP|9758068|dbj frnE protein-like {Ara... 284 2e-77
TC80020 weakly similar to GP|9759350|dbj|BAB10005.1 WD-repeat pr... 28 3.5
TC81100 similar to PIR|H84824|H84824 En/Spm-like transposon prot... 28 3.5
>TC87423 weakly similar to GP|9758068|dbj|BAB08647.1 frnE protein-like
{Arabidopsis thaliana}, partial (94%)
Length = 1169
Score = 439 bits (1130), Expect = e-124
Identities = 217/217 (100%), Positives = 217/217 (100%)
Frame = +1
Query: 1 MTDTNSAHSEKKLVRIDISSDTVCPWCFVGKKNLDKAIASSKDKYNFEILWHPYQLSPDA 60
MTDTNSAHSEKKLVRIDISSDTVCPWCFVGKKNLDKAIASSKDKYNFEILWHPYQLSPDA
Sbjct: 181 MTDTNSAHSEKKLVRIDISSDTVCPWCFVGKKNLDKAIASSKDKYNFEILWHPYQLSPDA 360
Query: 61 PKEGIEKREFYRSKFGSRSDQMEARMSEVFRTVGLVYSLSGLTGNTMDSHRLIYFSGQQG 120
PKEGIEKREFYRSKFGSRSDQMEARMSEVFRTVGLVYSLSGLTGNTMDSHRLIYFSGQQG
Sbjct: 361 PKEGIEKREFYRSKFGSRSDQMEARMSEVFRTVGLVYSLSGLTGNTMDSHRLIYFSGQQG 540
Query: 121 LDKQHDLVEELGLGYFTQEKYIGDQEFLLEAAAKVGIEGAEEFLKNPNNGLKEVEEELKT 180
LDKQHDLVEELGLGYFTQEKYIGDQEFLLEAAAKVGIEGAEEFLKNPNNGLKEVEEELKT
Sbjct: 541 LDKQHDLVEELGLGYFTQEKYIGDQEFLLEAAAKVGIEGAEEFLKNPNNGLKEVEEELKT 720
Query: 181 YSRNITGVPYYVINGSQKLSGGQPPEVFLRAFQAATS 217
YSRNITGVPYYVINGSQKLSGGQPPEVFLRAFQAATS
Sbjct: 721 YSRNITGVPYYVINGSQKLSGGQPPEVFLRAFQAATS 831
>CB892641 weakly similar to GP|9758068|dbj frnE protein-like {Arabidopsis
thaliana}, partial (87%)
Length = 778
Score = 390 bits (1001), Expect = e-109
Identities = 196/209 (93%), Positives = 200/209 (94%)
Frame = +2
Query: 9 SEKKLVRIDISSDTVCPWCFVGKKNLDKAIASSKDKYNFEILWHPYQLSPDAPKEGIEKR 68
SEKKLVRIDISSDTVCPWCFVGKKNLDKAIASSKDKYNFEILWHPYQLSPDAPKEGIEKR
Sbjct: 2 SEKKLVRIDISSDTVCPWCFVGKKNLDKAIASSKDKYNFEILWHPYQLSPDAPKEGIEKR 181
Query: 69 EFYRSKFGSRSDQMEARMSEVFRTVGLVYSLSGLTGNTMDSHRLIYFSGQQGLDKQHDLV 128
EFYRSKFGSRSDQMEARMSE R+ L + + + GNTMDSHRLIYFSGQQGLDKQHDLV
Sbjct: 182 EFYRSKFGSRSDQMEARMSEDSRSC-L*FVRTHVRGNTMDSHRLIYFSGQQGLDKQHDLV 358
Query: 129 EELGLGYFTQEKYIGDQEFLLEAAAKVGIEGAEEFLKNPNNGLKEVEEELKTYSRNITGV 188
EELGLGYFTQEKYIGDQEFLLEAAAKVGIEGAEEFLKNPNNGLKEVEEELKTYSRNITGV
Sbjct: 359 EELGLGYFTQEKYIGDQEFLLEAAAKVGIEGAEEFLKNPNNGLKEVEEELKTYSRNITGV 538
Query: 189 PYYVINGSQKLSGGQPPEVFLRAFQAATS 217
PYYVINGSQKLSGGQPPEVFLRAFQAATS
Sbjct: 539 PYYVINGSQKLSGGQPPEVFLRAFQAATS 625
>BF644778 weakly similar to GP|9758068|dbj frnE protein-like {Arabidopsis
thaliana}, partial (58%)
Length = 520
Score = 284 bits (727), Expect = 2e-77
Identities = 138/139 (99%), Positives = 138/139 (99%)
Frame = +2
Query: 1 MTDTNSAHSEKKLVRIDISSDTVCPWCFVGKKNLDKAIASSKDKYNFEILWHPYQLSPDA 60
MTDTNSAHSEKKLVRIDISSDTVCPWCFVGKKNLDKAIASSKDKYNFEILWHPYQLSPDA
Sbjct: 104 MTDTNSAHSEKKLVRIDISSDTVCPWCFVGKKNLDKAIASSKDKYNFEILWHPYQLSPDA 283
Query: 61 PKEGIEKREFYRSKFGSRSDQMEARMSEVFRTVGLVYSLSGLTGNTMDSHRLIYFSGQQG 120
PKEGIEKREFYRSKFGSRSDQMEARMSEVFRTVGLVYSLSGLTGNTMDSHRLIYFSGQQG
Sbjct: 284 PKEGIEKREFYRSKFGSRSDQMEARMSEVFRTVGLVYSLSGLTGNTMDSHRLIYFSGQQG 463
Query: 121 LDKQHDLVEELGLGYFTQE 139
LDKQHDLVEELGLGYFT E
Sbjct: 464 LDKQHDLVEELGLGYFTXE 520
>TC80020 weakly similar to GP|9759350|dbj|BAB10005.1 WD-repeat protein-like
{Arabidopsis thaliana}, partial (13%)
Length = 706
Score = 27.7 bits (60), Expect = 3.5
Identities = 12/24 (50%), Positives = 14/24 (58%)
Frame = -2
Query: 36 KAIASSKDKYNFEILWHPYQLSPD 59
KAI SS D ++WHP L PD
Sbjct: 156 KAI*SSPDSDERPVIWHPRPLKPD 85
>TC81100 similar to PIR|H84824|H84824 En/Spm-like transposon protein
[imported] - Arabidopsis thaliana, partial (20%)
Length = 1266
Score = 27.7 bits (60), Expect = 3.5
Identities = 18/54 (33%), Positives = 28/54 (51%), Gaps = 2/54 (3%)
Frame = -3
Query: 128 VEELGLGYFTQEKYIGDQEFLLEA--AAKVGIEGAEEFLKNPNNGLKEVEEELK 179
VE L +G + G +E A +VG+EG E+F+ N GL + E+L+
Sbjct: 910 VEALEVGLVGVDDLTGGAVDFVEVNVAREVGVEGLEDFVVEGNVGLPDGVEDLE 749
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.314 0.134 0.384
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,898,651
Number of Sequences: 36976
Number of extensions: 68799
Number of successful extensions: 274
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 273
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 273
length of query: 217
length of database: 9,014,727
effective HSP length: 92
effective length of query: 125
effective length of database: 5,612,935
effective search space: 701616875
effective search space used: 701616875
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 56 (26.2 bits)
Medicago: description of AC137823.6


