

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137823.1 + phase: 0
(249 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
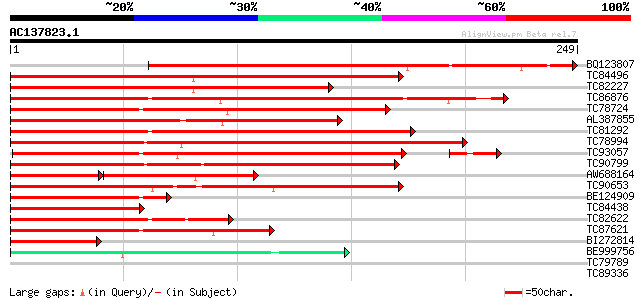
Score E
Sequences producing significant alignments: (bits) Value
BQ123807 similar to GP|13810202|em MADS2 protein {Cucumis sativu... 310 4e-85
TC84496 similar to GP|1483232|emb|CAA67969.1 MADS5 protein {Betu... 184 4e-47
TC82227 similar to GP|1483232|emb|CAA67969.1 MADS5 protein {Betu... 166 1e-41
TC86876 similar to GP|16973296|emb|CAC80857. C-type MADS box pro... 157 3e-39
TC78724 similar to GP|4322475|gb|AAD16052.1| putative MADS box t... 157 5e-39
AL387855 similar to GP|1483230|emb MADS4 protein {Betula pendula... 151 2e-37
TC81292 homologue to PIR|T09700|T09700 MADS-box protein - alfalf... 149 1e-36
TC78994 similar to SP|O64645|SOC1_ARATH SUPPRESSOR OF CONSTANS O... 147 5e-36
TC93057 homologue to GP|23194453|gb|AAN15183.1 MADS box protein ... 143 4e-35
TC90799 similar to GP|7672991|gb|AAF66690.1| MADS-box transcript... 120 4e-28
AW688164 similar to GP|3114588|gb|A MADS box protein {Eucalyptus... 73 3e-27
TC90653 similar to GP|10835358|gb|AAC13695.2 PTD protein {Populu... 114 3e-26
BE124909 similar to GP|4322475|gb| putative MADS box transcripti... 106 7e-24
TC84438 homologue to GP|1483232|emb|CAA67969.1 MADS5 protein {Be... 102 1e-22
TC82622 similar to GP|21739238|emb|CAD40993. OSJNBa0072F16.17 {O... 95 2e-20
TC87621 similar to SP|Q9FVC1|SVP_ARATH SHORT VEGETATIVE PHASE pr... 92 2e-19
BI272814 PIR|S71757|S71 MADS box protein DEFH200 - garden snapdr... 82 2e-16
BE999756 weakly similar to GP|9758603|dbj| gene_id:MDF20.13~unkn... 44 4e-05
TC79789 similar to PIR|D96519|D96519 mysoin-like protein 11013-... 32 0.30
TC89336 weakly similar to GP|21554135|gb|AAM63215.1 unknown {Ara... 32 0.30
>BQ123807 similar to GP|13810202|em MADS2 protein {Cucumis sativus}, partial
(88%)
Length = 676
Score = 310 bits (793), Expect = 4e-85
Identities = 162/191 (84%), Positives = 174/191 (90%), Gaps = 3/191 (1%)
Frame = +3
Query: 62 NMLKTLDRYQKCSYGAVEVSKPAKELESSYREYLKLKQRFENLQRAQRNLLGEDLGPLSS 121
+MLKTL RYQKCSYGAVEV+KPAKELESSYREYLKLK RFE+LQR QRNLLGEDLGPL +
Sbjct: 6 SMLKTLVRYQKCSYGAVEVNKPAKELESSYREYLKLKARFESLQRTQRNLLGEDLGPLGT 185
Query: 122 KDLEQLERQLDSSLKQVRSTKTQFMLDQLADLQNKEHMLVEANRSLSIKLEE--INSRNH 179
KDLEQLERQLDSSLKQVRSTKTQFMLDQLADLQNKEHMLVEANRSLS+KLEE INSRN
Sbjct: 186 KDLEQLERQLDSSLKQVRSTKTQFMLDQLADLQNKEHMLVEANRSLSMKLEEININSRNQ 365
Query: 180 YRQSWEASDQSMQYEAQQNAHSQSFFQQLECNPTLQIGSDYRYN-NVASDQIASTSQAQQ 238
YRQ+WEA DQSM Y QNAHSQSFFQ LECNPTLQIG+DYRY+ VASDQ+ +T+QA Q
Sbjct: 366 YRQTWEAGDQSMAY-GNQNAHSQSFFQPLECNPTLQIGTDYRYSPPVASDQLTATTQA-Q 539
Query: 239 QVNGFVPGWML 249
QVNGF+PGWML
Sbjct: 540 QVNGFIPGWML 572
>TC84496 similar to GP|1483232|emb|CAA67969.1 MADS5 protein {Betula
pendula}, partial (74%)
Length = 681
Score = 184 bits (466), Expect = 4e-47
Identities = 95/174 (54%), Positives = 130/174 (74%), Gaps = 1/174 (0%)
Frame = +2
Query: 1 MGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVALIIFSTRGKLYEFCST 60
MGRGRV+LK+IENKINRQVTF+KRR+GLLKKA+E+SVLCDAEVALI+FS +GKLYE+ S
Sbjct: 26 MGRGRVQLKKIENKINRQVTFSKRRSGLLKKAHEISVLCDAEVALIVFSNKGKLYEYSSD 205
Query: 61 SNMLKTLDRYQKCSYGAVE-VSKPAKELESSYREYLKLKQRFENLQRAQRNLLGEDLGPL 119
M K L+RY++ SY + V+ + E+ E+ +LK R E +Q+ QRN +GE+L L
Sbjct: 206 PCMEKILERYERYSYAERQHVANDQPQNENWIIEHARLKTRLEVIQKNQRNFMGEELDGL 385
Query: 120 SSKDLEQLERQLDSSLKQVRSTKTQFMLDQLADLQNKEHMLVEANRSLSIKLEE 173
S K+L+ LE QLDS+LKQ+RS K Q M + +++L K+ L E N+ L+ K++E
Sbjct: 386 SMKELQHLEHQLDSALKQIRSRKNQLMYESISELSKKDKALQEKNKLLTTKMKE 547
>TC82227 similar to GP|1483232|emb|CAA67969.1 MADS5 protein {Betula
pendula}, partial (68%)
Length = 681
Score = 166 bits (419), Expect = 1e-41
Identities = 89/143 (62%), Positives = 110/143 (76%), Gaps = 1/143 (0%)
Frame = +1
Query: 1 MGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVALIIFSTRGKLYEFCST 60
MGRGRV+LKRIENKINRQVTF+KRR+GLLKKA E+SVLCDAEVALIIFST+GKL+E+ S
Sbjct: 130 MGRGRVQLKRIENKINRQVTFSKRRSGLLKKAQEISVLCDAEVALIIFSTKGKLFEYSSD 309
Query: 61 SNMLKTLDRYQKCSYGAVE-VSKPAKELESSYREYLKLKQRFENLQRAQRNLLGEDLGPL 119
M K L+RY++CSY + V+ E+ E+ KLK R E LQR QRN +GEDL L
Sbjct: 310 PCMEKILERYERCSYMERQLVTSEQSPNENWVLEHAKLKARMEVLQRNQRNFMGEDLDGL 489
Query: 120 SSKDLEQLERQLDSSLKQVRSTK 142
K+L+ LE+QLDS+LKQ+ T+
Sbjct: 490 GLKELQSLEQQLDSALKQITITE 558
>TC86876 similar to GP|16973296|emb|CAC80857. C-type MADS box protein {Malus
x domestica}, partial (85%)
Length = 2050
Score = 157 bits (398), Expect = 3e-39
Identities = 95/222 (42%), Positives = 136/222 (60%), Gaps = 3/222 (1%)
Frame = +2
Query: 1 MGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVALIIFSTRGKLYEFCST 60
MGRG++E+KRIEN NRQVTF KRRNGLLKKAYELSVLCDAEVAL++FSTRG+LYE+ +
Sbjct: 89 MGRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALVVFSTRGRLYEYANN 268
Query: 61 SNMLKTLDRYQKCSYGAVEVSKPAKELESSY-REYLKLKQRFENLQRAQRNLLGEDLGPL 119
S + T++RY+K + ++ Y +E KL+++ ++Q R++LGE LG L
Sbjct: 269 S-VRATIERYKKACAASTNAESVSEANTQFYQQESSKLRRQIRDIQNLNRHILGEALGSL 445
Query: 120 SSKDLEQLERQLDSSLKQVRSTKTQFMLDQLADLQNKEHMLVEANRSLSIKLEEINSRNH 179
S K+L+ LE +L+ L +VRS K + + + +Q +E L N L K+ E + R
Sbjct: 446 SLKELKNLEGRLEKGLSRVRSRKHETLFADVEFMQKREIELQNHNNYLRAKIAE-HERAQ 622
Query: 180 YRQSWEASDQSM--QYEAQQNAHSQSFFQQLECNPTLQIGSD 219
+Q DQ+M Q A+ ++FF P +GSD
Sbjct: 623 QQQHNLMPDQTMCDQSLPSSQAYDRNFF------PVNLLGSD 730
>TC78724 similar to GP|4322475|gb|AAD16052.1| putative MADS box
transcription factor ETL {Eucalyptus globulus subsp.
globulus}, partial (71%)
Length = 1089
Score = 157 bits (396), Expect = 5e-39
Identities = 84/168 (50%), Positives = 123/168 (73%), Gaps = 1/168 (0%)
Frame = +1
Query: 1 MGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVALIIFSTRGKLYEFCST 60
M RG+ ++KRIEN+ NRQVTF+KRRNGLLKKA+ELSVLCDAEVALI+FST GKLYEF S+
Sbjct: 226 MVRGKTQMKRIENESNRQVTFSKRRNGLLKKAFELSVLCDAEVALIVFSTTGKLYEF-SS 402
Query: 61 SNMLKTLDRYQKCSYGAVEVSKPAKELESSYREY-LKLKQRFENLQRAQRNLLGEDLGPL 119
S++ KT++RYQ V +K +E +E + ++ E+L+ ++R LLGE+LG
Sbjct: 403 SSISKTVERYQGKVKELVLSTKGIQENTQHLKECDIDTTKKLEHLELSKRKLLGEELGSC 582
Query: 120 SSKDLEQLERQLDSSLKQVRSTKTQFMLDQLADLQNKEHMLVEANRSL 167
+ +L+Q+E QL+ SL ++R+ K Q + +Q+ L++KE +L+E N+ L
Sbjct: 583 AFDELQQIENQLERSLSKIRARKNQLLKEQIEKLKDKERLLLEENKRL 726
>AL387855 similar to GP|1483230|emb MADS4 protein {Betula pendula}, partial
(51%)
Length = 493
Score = 151 bits (382), Expect = 2e-37
Identities = 79/147 (53%), Positives = 111/147 (74%), Gaps = 1/147 (0%)
Frame = +2
Query: 1 MGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVALIIFSTRGKLYEFCST 60
MGRGRV+LKRIENK ++QVTF KRR GLLKKA E+SVLCDA+VALI+FST+GKL+E+ S
Sbjct: 53 MGRGRVQLKRIENKTSQQVTFFKRRTGLLKKANEISVLCDAQVALIMFSTKGKLFEYSSA 232
Query: 61 SNMLKTLDRYQKCSYGAVEVSKPAKELESSYR-EYLKLKQRFENLQRAQRNLLGEDLGPL 119
+M L+RY++ ++ E++ E + ++ EY+KL + + L+R RN +G DL PL
Sbjct: 233 PSMEDILERYERQNH--TELTGATNETQGNWSFEYMKLTAKVQVLERNLRNFVGNDLDPL 406
Query: 120 SSKDLEQLERQLDSSLKQVRSTKTQFM 146
S K+L+ LE+QLD+SLK++R+ K Q M
Sbjct: 407 SVKELQSLEQQLDTSLKRIRTRKNQVM 487
>TC81292 homologue to PIR|T09700|T09700 MADS-box protein - alfalfa
(fragment), partial (91%)
Length = 800
Score = 149 bits (376), Expect = 1e-36
Identities = 77/178 (43%), Positives = 116/178 (64%)
Frame = +2
Query: 1 MGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVALIIFSTRGKLYEFCST 60
MGRG+++++RI+N +RQVTF+KRRNGLLKKA EL++LCDAEV ++IFS+ KLY+F ST
Sbjct: 71 MGRGKIQIRRIDNSTSRQVTFSKRRNGLLKKAKELAILCDAEVGVMIFSSTAKLYDFAST 250
Query: 61 SNMLKTLDRYQKCSYGAVEVSKPAKELESSYREYLKLKQRFENLQRAQRNLLGEDLGPLS 120
S M +DRY K ++ E++ RE L+Q+ NLQ + R ++GE+L L+
Sbjct: 251 S-MRSVIDRYNKTKEEHNQLGSSTSEIKFWQREAAMLRQQLHNLQESHRQIMGEELSGLT 427
Query: 121 SKDLEQLERQLDSSLKQVRSTKTQFMLDQLADLQNKEHMLVEANRSLSIKLEEINSRN 178
K+L+ LE QL+ SL+ VR K Q +D++ +L K ++ + N L K+ +N
Sbjct: 428 VKELQGLENQLEISLRGVRMKKEQLFMDEIQELNRKGDIIHQENVELYRKVYGTKDKN 601
>TC78994 similar to SP|O64645|SOC1_ARATH SUPPRESSOR OF CONSTANS
OVEREXPRESSION 1 protein (Agamous-like MADS box protein
AGL20)., partial (83%)
Length = 946
Score = 147 bits (370), Expect = 5e-36
Identities = 83/203 (40%), Positives = 128/203 (62%), Gaps = 2/203 (0%)
Frame = +3
Query: 1 MGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVALIIFSTRGKLYEFCST 60
M RG+ ++KRIEN +RQVTF+KRRNGLLKKA+ELSVLCDAEVALI+FS RG+LYEF S
Sbjct: 54 MVRGKTQMKRIENATSRQVTFSKRRNGLLKKAFELSVLCDAEVALIVFSPRGRLYEFAS- 230
Query: 61 SNMLKTLDRYQKCS--YGAVEVSKPAKELESSYREYLKLKQRFENLQRAQRNLLGEDLGP 118
S++L+T++RY+ + S+ + + E + ++ + L+ ++R LLGE LG
Sbjct: 231 SSILETIERYRSHTRINNTPTTSESVENTQQLKEEAENMMKKIDLLETSKRKLLGEGLGS 410
Query: 119 LSSKDLEQLERQLDSSLKQVRSTKTQFMLDQLADLQNKEHMLVEANRSLSIKLEEINSRN 178
S +L+++E+QL+ S+ ++R KT+ +Q+ L+ KE LV N LS K +++
Sbjct: 411 CSIDELQKIEQQLEKSINKIRVKKTKVFREQIDQLKEKEKALVAENVRLSEKYGNYSTQE 590
Query: 179 HYRQSWEASDQSMQYEAQQNAHS 201
+ E ++ Y Q + S
Sbjct: 591 STKDQRENIAEAEPYADQSSPSS 659
>TC93057 homologue to GP|23194453|gb|AAN15183.1 MADS box protein GHMADS-2
{Gossypium hirsutum}, partial (95%)
Length = 858
Score = 143 bits (361), Expect(2) = 4e-35
Identities = 79/174 (45%), Positives = 115/174 (65%), Gaps = 1/174 (0%)
Frame = +3
Query: 2 GRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVALIIFSTRGKLYEFCSTS 61
G R+ K IEN NRQVTF KRRNGLLKKAYELSVLCDAEVALI+FS+RG+LYE+ S +
Sbjct: 33 GGERLR*KGIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSSRGRLYEY-SNN 209
Query: 62 NMLKTLDRYQK-CSYGAVEVSKPAKELESSYREYLKLKQRFENLQRAQRNLLGEDLGPLS 120
N+ T+DRY+K CS + + + +E KL+Q+ + LQ + R+L+G+ L L+
Sbjct: 210 NIRSTIDRYKKACSDHSSTTTTTEINAQYYQQESAKLRQQIQMLQNSNRHLMGDALSTLT 389
Query: 121 SKDLEQLERQLDSSLKQVRSTKTQFMLDQLADLQNKEHMLVEANRSLSIKLEEI 174
K+L+QLE +L+ + ++RS K + +L ++ Q +E L N L K+ ++
Sbjct: 390 VKELKQLENRLERGITRIRSKKHEMLLAEIEYFQKREIELENENLCLRTKINDV 551
Score = 21.6 bits (44), Expect(2) = 4e-35
Identities = 9/23 (39%), Positives = 14/23 (60%)
Frame = +2
Query: 194 EAQQNAHSQSFFQQLECNPTLQI 216
EA + H F+ + ECNP++ I
Sbjct: 554 EASSSEHG--FWTRAECNPSISI 616
>TC90799 similar to GP|7672991|gb|AAF66690.1| MADS-box transcription factor
{Canavalia lineata}, partial (76%)
Length = 653
Score = 120 bits (302), Expect = 4e-28
Identities = 69/171 (40%), Positives = 107/171 (62%)
Frame = +2
Query: 1 MGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVALIIFSTRGKLYEFCST 60
M R ++++K+I+N RQVTF+KRR G+ KKA ELS+LCDAEV L+IFST GKLYE+ S
Sbjct: 71 MARQKIKIKKIDNATARQVTFSKRRRGIFKKAEELSILCDAEVGLVIFSTTGKLYEYAS- 247
Query: 61 SNMLKTLDRYQKCSYGAVEVSKPAKELESSYREYLKLKQRFENLQRAQRNLLGEDLGPLS 120
SNM + RY + S+ ++ KP +++ +L + + + R + ED L+
Sbjct: 248 SNMKDIITRYGQQSHHITKLDKPL-QVQVEKNMPAELNKEVADRTQQLRGMKSEDFEGLN 424
Query: 121 SKDLEQLERQLDSSLKQVRSTKTQFMLDQLADLQNKEHMLVEANRSLSIKL 171
+ L+QLE+ L+S LK+V K + +L+++ L+ KE ML N+ L K+
Sbjct: 425 LEGLQQLEKSLESXLKRVIEMKEKKILNEIKALRMKEIMLEXENKHLKQKM 577
>AW688164 similar to GP|3114588|gb|A MADS box protein {Eucalyptus grandis},
partial (31%)
Length = 549
Score = 72.8 bits (177), Expect(2) = 3e-27
Identities = 35/41 (85%), Positives = 39/41 (94%)
Frame = +1
Query: 1 MGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDA 41
MGRGRV L+RIENKINRQVTF+KRR+GLLKKA+EL VLCDA
Sbjct: 7 MGRGRVVLERIENKINRQVTFSKRRSGLLKKAFELCVLCDA 129
Score = 66.2 bits (160), Expect(2) = 3e-27
Identities = 32/70 (45%), Positives = 51/70 (72%), Gaps = 2/70 (2%)
Frame = +2
Query: 42 EVALIIFSTRGKLYEFCSTSNMLKTLDRYQKCSYGAVEV--SKPAKELESSYREYLKLKQ 99
EVALIIFS+RGKL+++ ST+++ K ++RY++C Y + S E ++ Y++YLKLK
Sbjct: 131 EVALIIFSSRGKLFQYSSTTDINKIIERYRQCRYSKPQAGNSLGHNESQNLYQDYLKLKA 310
Query: 100 RFENLQRAQR 109
++E+L R QR
Sbjct: 311 KYESLDRKQR 340
>TC90653 similar to GP|10835358|gb|AAC13695.2 PTD protein {Populus
balsamifera subsp. trichocarpa}, partial (59%)
Length = 845
Score = 114 bits (286), Expect = 3e-26
Identities = 68/177 (38%), Positives = 108/177 (60%), Gaps = 4/177 (2%)
Frame = +2
Query: 1 MGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVALIIFSTRGKLYEFCST 60
MGRG++E+K IEN NRQVT++KRRNG+ KKA+ELSVLCDA+V+LI+FS K++E+ +
Sbjct: 44 MGRGKIEIKLIENPTNRQVTYSKRRNGIFKKAHELSVLCDAKVSLIMFSKNNKMHEYITP 223
Query: 61 S-NMLKTLDRYQKCSYGAVEVSKPAKELESSYREYLKLKQRFENLQRAQRNLLGE---DL 116
+ K +D+YQK + G +++ + E KLK L+R R+ +GE +L
Sbjct: 224 GLSTKKIIDQYQK-TLGDIDLWR--SHYEKMLENLKKLKDINHKLRRQIRHRIGEGGMEL 394
Query: 117 GPLSSKDLEQLERQLDSSLKQVRSTKTQFMLDQLADLQNKEHMLVEANRSLSIKLEE 173
LS + L LE ++SS+ ++R K + + + K L + N +L ++LE+
Sbjct: 395 DDLSFQQLRSLEEDMNSSIAKIRERKFHVIKTRTDTCRKKVRSLEQMNGNLLLELEK 565
>BE124909 similar to GP|4322475|gb| putative MADS box transcription factor
ETL {Eucalyptus globulus subsp. globulus}, partial (60%)
Length = 608
Score = 106 bits (265), Expect = 7e-24
Identities = 53/71 (74%), Positives = 65/71 (90%)
Frame = +1
Query: 1 MGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVALIIFSTRGKLYEFCST 60
M RG+ ++KRIEN+ NRQVTF+KRRNGLLKKA+ELSVLCDAEVALI+FST GKLYEF S+
Sbjct: 148 MVRGKTQMKRIENESNRQVTFSKRRNGLLKKAFELSVLCDAEVALIVFSTTGKLYEF-SS 324
Query: 61 SNMLKTLDRYQ 71
S++ KT++RYQ
Sbjct: 325 SSISKTVERYQ 357
>TC84438 homologue to GP|1483232|emb|CAA67969.1 MADS5 protein {Betula
pendula}, partial (24%)
Length = 702
Score = 102 bits (255), Expect = 1e-22
Identities = 50/59 (84%), Positives = 57/59 (95%)
Frame = +1
Query: 1 MGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVALIIFSTRGKLYEFCS 59
MGRGRV+LKRIENKINRQVTF+KRR+GLLKKA E+SVLCDAEVALIIFST+GKL+E+ S
Sbjct: 223 MGRGRVQLKRIENKINRQVTFSKRRSGLLKKAQEISVLCDAEVALIIFSTKGKLFEYSS 399
>TC82622 similar to GP|21739238|emb|CAD40993. OSJNBa0072F16.17 {Oryza
sativa}, partial (78%)
Length = 847
Score = 95.1 bits (235), Expect = 2e-20
Identities = 48/98 (48%), Positives = 70/98 (70%)
Frame = +3
Query: 1 MGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVALIIFSTRGKLYEFCST 60
MGRG++ ++RI+N +RQVTF+KRR GL+KKA EL++LCDA+V L+IFS+ GKLYE+ +T
Sbjct: 138 MGRGKIVIRRIDNCTSRQVTFSKRRKGLIKKAKELAILCDAQVGLVIFSSTGKLYEYANT 317
Query: 61 SNMLKTLDRYQKCSYGAVEVSKPAKELESSYREYLKLK 98
S M ++RY C +V+ P E++ RE + K
Sbjct: 318 S-MKSVIERYNICKEDQ-QVTNPESEVKFWQREAVHFK 425
>TC87621 similar to SP|Q9FVC1|SVP_ARATH SHORT VEGETATIVE PHASE protein.
[Mouse-ear cress] {Arabidopsis thaliana}, partial (65%)
Length = 1322
Score = 92.0 bits (227), Expect = 2e-19
Identities = 52/117 (44%), Positives = 75/117 (63%), Gaps = 1/117 (0%)
Frame = +2
Query: 1 MGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVALIIFSTRGKLYEFCST 60
M R ++++K+IEN RQVTF+KRR GL+KKA ELSVLCDA+VALIIFS+ GKL+E+ S
Sbjct: 437 MAREKIQIKKIENSTARQVTFSKRRRGLIKKAEELSVLCDADVALIIFSSTGKLFEY-SN 613
Query: 61 SNMLKTLDRYQKCSYGAVEVSKPAKELE-SSYREYLKLKQRFENLQRAQRNLLGEDL 116
+M + L+R+ S ++ +P+ EL+ +L + R + GEDL
Sbjct: 614 LSMREILERHHLHSKNLAKLEEPSLELQLVENSNCSRLSKEVAQKSHQLRQMRGEDL 784
>BI272814 PIR|S71757|S71 MADS box protein DEFH200 - garden snapdragon,
partial (16%)
Length = 320
Score = 82.0 bits (201), Expect = 2e-16
Identities = 40/40 (100%), Positives = 40/40 (100%)
Frame = +1
Query: 1 MGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCD 40
MGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCD
Sbjct: 199 MGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCD 318
>BE999756 weakly similar to GP|9758603|dbj| gene_id:MDF20.13~unknown protein
{Arabidopsis thaliana}, partial (15%)
Length = 534
Score = 44.3 bits (103), Expect = 4e-05
Identities = 36/163 (22%), Positives = 63/163 (38%), Gaps = 14/163 (8%)
Frame = +1
Query: 1 MGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVALIIF------------ 48
M +V+L I N R+ T+ KR L+KK E++ LC + I+F
Sbjct: 13 MAGNKVKLAFITNHTARRATYKKRVQSLMKKLNEITTLCGVKACGIVFKPDDLEPQIWPS 192
Query: 49 --STRGKLYEFCSTSNMLKTLDRYQKCSYGAVEVSKPAKELESSYREYLKLKQRFENLQR 106
L F T N + + SY + K ++L+ +E + +
Sbjct: 193 IEGVHSVLVRFMQTPNFYRDRKMFDHESYLKERIQKLNEKLKKKMKENRMMWMSVQLHHY 372
Query: 107 AQRNLLGEDLGPLSSKDLEQLERQLDSSLKQVRSTKTQFMLDQ 149
+ + ED LS+ D+ L +D +K++ Q D+
Sbjct: 373 LEAGNVPED---LSTSDMNDLTYVVDEKMKEINMKMVQLEKDE 492
>TC79789 similar to PIR|D96519|D96519 mysoin-like protein 11013-7318
[imported] - Arabidopsis thaliana, partial (21%)
Length = 1097
Score = 31.6 bits (70), Expect = 0.30
Identities = 24/114 (21%), Positives = 55/114 (48%), Gaps = 3/114 (2%)
Frame = +3
Query: 84 AKELESSYREYLKLKQRFENLQRAQRNLLGEDLGPLSSKDLEQLERQLDSSLKQVRSTKT 143
A +L+ + +E ++ + NL+ + L +D+ +K L++++ L++ ++
Sbjct: 630 ASQLDGALKECMR---QIRNLKE-EHELKIQDISLAKTKQLDKIKGDLEARIRNFEQELL 797
Query: 144 QFMLDQLA---DLQNKEHMLVEANRSLSIKLEEINSRNHYRQSWEASDQSMQYE 194
+ D A LQ + +MLV+ + + EI + + +S E S++YE
Sbjct: 798 RSAADNAALSRSLQERSNMLVKLSEEKAHAEAEIEHQKNTVESCEREINSLKYE 959
>TC89336 weakly similar to GP|21554135|gb|AAM63215.1 unknown {Arabidopsis
thaliana}, partial (7%)
Length = 1241
Score = 31.6 bits (70), Expect = 0.30
Identities = 33/128 (25%), Positives = 55/128 (42%), Gaps = 27/128 (21%)
Frame = +1
Query: 79 EVSKPAKELESSYREYLKLKQRFENLQRAQRNLLGEDLGPLSSKDLEQLERQLDSSLKQV 138
EV++ KELES+ E +L+ + E ++ +++ +LE QLDS KQ+
Sbjct: 754 EVARLRKELESNLAETRQLQDQLE----------------VAQENVTKLEWQLDSGRKQI 885
Query: 139 R----------------STKTQFMLDQLADLQ-----NKEHM------LVEANRSLSIKL 171
R + Q + D++ D+Q K+ + L E L+ L
Sbjct: 886 RELEDRITWFKTNETNLEVEVQKLKDEMHDVQAQFSFEKDQLHSDIASLSEIKTQLTSTL 1065
Query: 172 EEINSRNH 179
EE SR+H
Sbjct: 1066EEWESRSH 1089
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.315 0.129 0.353
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,199,415
Number of Sequences: 36976
Number of extensions: 69746
Number of successful extensions: 364
Number of sequences better than 10.0: 52
Number of HSP's better than 10.0 without gapping: 350
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 352
length of query: 249
length of database: 9,014,727
effective HSP length: 94
effective length of query: 155
effective length of database: 5,538,983
effective search space: 858542365
effective search space used: 858542365
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 57 (26.6 bits)
Medicago: description of AC137823.1


