

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137822.17 - phase: 0
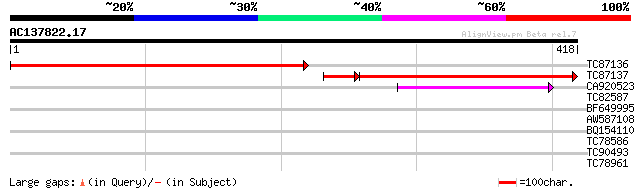
(418 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC87136 similar to GP|15292751|gb|AAK92744.1 unknown protein {Ar... 436 e-123
TC87137 similar to GP|15292751|gb|AAK92744.1 unknown protein {Ar... 322 2e-98
CA920523 similar to PIR|H86313|H86 protein F2H15.10 [imported] -... 79 3e-15
TC82587 similar to PIR|H86313|H86313 protein F2H15.10 [imported]... 37 0.018
BF649995 similar to PIR|H86313|H863 protein F2H15.10 [imported] ... 36 0.024
AW587108 similar to PIR|H86313|H863 protein F2H15.10 [imported] ... 36 0.032
BQ154110 similar to GP|17065220|gb| RNA-binding protein-like {Ar... 29 2.9
TC78586 similar to PIR|T01041|T01041 hypothetical protein YUP8H1... 29 3.9
TC90493 similar to GP|13937218|gb|AAK50101.1 AT5g17630/K10A8_110... 28 5.0
TC78961 similar to GP|18252179|gb|AAL61922.1 unknown protein {Ar... 28 5.0
>TC87136 similar to GP|15292751|gb|AAK92744.1 unknown protein {Arabidopsis
thaliana}, partial (41%)
Length = 1167
Score = 436 bits (1122), Expect = e-123
Identities = 218/220 (99%), Positives = 218/220 (99%)
Frame = +1
Query: 1 MELLGPEKVDPADVKKIKDKLFGYSTFWVTKEEPFGELGEGILFIGNLRGKREDIFSILQ 60
MELLGPEKVDPADVKKIKDKLFGYSTFWVTKEEPFGELGEGILFIGNLRGKREDIFSILQ
Sbjct: 508 MELLGPEKVDPADVKKIKDKLFGYSTFWVTKEEPFGELGEGILFIGNLRGKREDIFSILQ 687
Query: 61 NRLVEATGDKYNLFMVEEPDSDSPDPRGGPRVSFGLLRKEVSEPEETTLWQYVVASLLFL 120
NRLVEATGDKYNLFMVEEPDSDSPDPRGGPRVSFGLLRKEVSEPEETTLWQYV ASLL L
Sbjct: 688 NRLVEATGDKYNLFMVEEPDSDSPDPRGGPRVSFGLLRKEVSEPEETTLWQYVDASLLIL 867
Query: 121 LTIGTSVEVGIASQINRLPPELVKFLTDPNYTEAPDMEMLYPFVESALPLAYGVLGVLLF 180
LTIGTSVEVGIASQINRLPPELVKFLTDPNYTEAPDMEMLYPFVESALPLAYGVLGVLLF
Sbjct: 868 LTIGTSVEVGIASQINRLPPELVKFLTDPNYTEAPDMEMLYPFVESALPLAYGVLGVLLF 1047
Query: 181 HEVGHFLAAFPKQVKLSIPFFIPHITLGSFGAITQFKSIL 220
HEVGHFLAAFPKQVKLSIPFFIPHITLGSFGAITQFKSIL
Sbjct: 1048HEVGHFLAAFPKQVKLSIPFFIPHITLGSFGAITQFKSIL 1167
>TC87137 similar to GP|15292751|gb|AAK92744.1 unknown protein {Arabidopsis
thaliana}, partial (33%)
Length = 746
Score = 322 bits (826), Expect(2) = 2e-98
Identities = 159/160 (99%), Positives = 160/160 (99%)
Frame = +2
Query: 259 GDLVQVPSMLFQGSLLLGLISRATLGYAAVHAATVPIHPLVIAGWCGLTIQAFNMLPVGC 318
GDLVQVPSMLFQGSLLLGLISRATLGYAAVHAATVPIHPLVIAGWCGLTIQAFNMLPVGC
Sbjct: 95 GDLVQVPSMLFQGSLLLGLISRATLGYAAVHAATVPIHPLVIAGWCGLTIQAFNMLPVGC 274
Query: 319 LDGGRSVQGAFGKGATMVFGLTTYTLLGLGVLGGPLSLAWGFFVIFSQRSPEKPCLNDVT 378
LDGGRSVQGAFGKGATMVFGLTTYTLLGLGVLGGPLSLAWGFFVIFSQRSPEKPCLNDVT
Sbjct: 275 LDGGRSVQGAFGKGATMVFGLTTYTLLGLGVLGGPLSLAWGFFVIFSQRSPEKPCLNDVT 454
Query: 379 EVGTWRQTFVGVAIFLAVLTLLPVWDELAEELGIGLVTTF 418
EVGTWRQTFVGVAIFLAVLTLLPVWDELA+ELGIGLVTTF
Sbjct: 455 EVGTWRQTFVGVAIFLAVLTLLPVWDELAKELGIGLVTTF 574
Score = 55.1 bits (131), Expect(2) = 2e-98
Identities = 27/27 (100%), Positives = 27/27 (100%)
Frame = +1
Query: 232 AGPFAGAVLSFSMFAVGLLLSSNPDVA 258
AGPFAGAVLSFSMFAVGLLLSSNPDVA
Sbjct: 13 AGPFAGAVLSFSMFAVGLLLSSNPDVA 93
>CA920523 similar to PIR|H86313|H86 protein F2H15.10 [imported] - Arabidopsis
thaliana, partial (25%)
Length = 714
Score = 79.0 bits (193), Expect = 3e-15
Identities = 43/116 (37%), Positives = 59/116 (50%), Gaps = 1/116 (0%)
Frame = -1
Query: 287 AVHAATVPIHPLVIAGWCGLTIQAFNMLPVGCLDGGRSVQGAFGKGATMVFGLTTYTLLG 346
AV P+ PL AG G+ + + N+LP G L+GGR Q FG+ + T LLG
Sbjct: 636 AVEGVEFPVDPLAFAGLLGMVVTSLNLLPCGRLEGGRIAQAMFGRSTATLLSFGTSLLLG 457
Query: 347 LGVLGGP-LSLAWGFFVIFSQRSPEKPCLNDVTEVGTWRQTFVGVAIFLAVLTLLP 401
+G L G L LAWG F F + E P +++T +G R + V + LTL P
Sbjct: 456 IGGLSGSVLCLAWGLFATFFRGGEEIPAKDEITPLGESRYAWGIVLGLICFLTLFP 289
>TC82587 similar to PIR|H86313|H86313 protein F2H15.10 [imported] -
Arabidopsis thaliana, partial (24%)
Length = 764
Score = 36.6 bits (83), Expect = 0.018
Identities = 21/64 (32%), Positives = 37/64 (57%)
Frame = +2
Query: 13 DVKKIKDKLFGYSTFWVTKEEPFGELGEGILFIGNLRGKREDIFSILQNRLVEATGDKYN 72
D+ K+K FG+ TF+ T FG+ G +FIGNLR +++ L+ +L +A+ +
Sbjct: 584 DLNKLKS-CFGFDTFFTTDVRRFGDGG---IFIGNLRRPIDEVIPKLEKKLSDASSREVV 751
Query: 73 LFMV 76
L+ +
Sbjct: 752 LWFM 763
>BF649995 similar to PIR|H86313|H863 protein F2H15.10 [imported] -
Arabidopsis thaliana, partial (9%)
Length = 540
Score = 36.2 bits (82), Expect = 0.024
Identities = 18/47 (38%), Positives = 28/47 (59%)
Frame = +3
Query: 22 FGYSTFWVTKEEPFGELGEGILFIGNLRGKREDIFSILQNRLVEATG 68
FG+ TF+ T FG+ G +FIGNLR +++ L+ +L +A G
Sbjct: 291 FGFDTFFTTNVRRFGDGG---IFIGNLRRPIDEVIPKLEKKLSDAAG 422
>AW587108 similar to PIR|H86313|H863 protein F2H15.10 [imported] -
Arabidopsis thaliana, partial (9%)
Length = 647
Score = 35.8 bits (81), Expect = 0.032
Identities = 18/47 (38%), Positives = 28/47 (59%)
Frame = +3
Query: 22 FGYSTFWVTKEEPFGELGEGILFIGNLRGKREDIFSILQNRLVEATG 68
FG+ TF+ T FG+ G +FIGNLR +++ L+ +L +A G
Sbjct: 327 FGFDTFFTTDVRRFGDGG---IFIGNLRRPIDEVIPKLEKKLSDAAG 458
>BQ154110 similar to GP|17065220|gb| RNA-binding protein-like {Arabidopsis
thaliana}, partial (12%)
Length = 621
Score = 29.3 bits (64), Expect = 2.9
Identities = 14/27 (51%), Positives = 17/27 (62%), Gaps = 3/27 (11%)
Frame = -2
Query: 181 HEVGHFLAAFPKQVKLSIPF---FIPH 204
H GHFL+AFP Q L IP+ +PH
Sbjct: 185 HHGGHFLSAFPSQ*ILQIPWMQGLLPH 105
>TC78586 similar to PIR|T01041|T01041 hypothetical protein YUP8H12R.24 -
Arabidopsis thaliana, partial (38%)
Length = 2831
Score = 28.9 bits (63), Expect = 3.9
Identities = 15/41 (36%), Positives = 20/41 (48%)
Frame = -1
Query: 199 PFFIPHITLGSFGAITQFKSILPDRSTQVDISLAGPFAGAV 239
P F+ ++L F A+ F L S+ S GPF GAV
Sbjct: 521 PSFLLFLSLALFSALVSFTKPLSSLSSSSVSSFLGPFTGAV 399
>TC90493 similar to GP|13937218|gb|AAK50101.1 AT5g17630/K10A8_110
{Arabidopsis thaliana}, partial (70%)
Length = 1081
Score = 28.5 bits (62), Expect = 5.0
Identities = 23/73 (31%), Positives = 32/73 (43%), Gaps = 11/73 (15%)
Frame = +1
Query: 160 LYPFVESALPLAYGVLGVLLFHEVGHFLA---------AFPKQVKLSIPFF--IPHITLG 208
L P + + P + +LG LFH +GH A +F +K + P F I LG
Sbjct: 322 LQPCPKISKPFIFALLGPALFHTIGHISACVSFSKVAVSFTHVIKSAEPVFSVIFSSVLG 501
Query: 209 SFGAITQFKSILP 221
I + SILP
Sbjct: 502 DRYPIQVWLSILP 540
>TC78961 similar to GP|18252179|gb|AAL61922.1 unknown protein {Arabidopsis
thaliana}, partial (71%)
Length = 974
Score = 28.5 bits (62), Expect = 5.0
Identities = 12/24 (50%), Positives = 14/24 (58%)
Frame = -3
Query: 84 PDPRGGPRVSFGLLRKEVSEPEET 107
PDP GGPR + L +E EP T
Sbjct: 354 PDPGGGPRPKYSLESRESREPRGT 283
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.323 0.143 0.429
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,125,895
Number of Sequences: 36976
Number of extensions: 164125
Number of successful extensions: 695
Number of sequences better than 10.0: 20
Number of HSP's better than 10.0 without gapping: 690
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 692
length of query: 418
length of database: 9,014,727
effective HSP length: 99
effective length of query: 319
effective length of database: 5,354,103
effective search space: 1707958857
effective search space used: 1707958857
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 60 (27.7 bits)
Medicago: description of AC137822.17


