

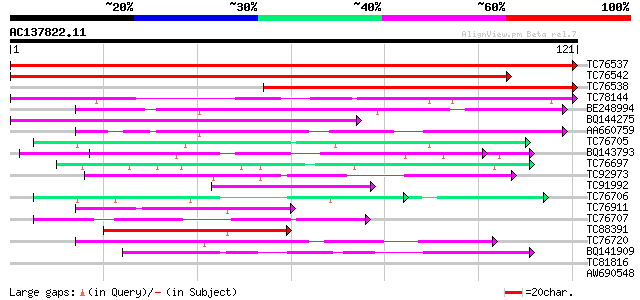
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137822.11 - phase: 0
(121 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC76537 homologue to GP|6969496|gb|AAF33785.1| cold acclimation ... 271 6e-74
TC76542 homologue to GP|6969496|gb|AAF33785.1| cold acclimation ... 220 9e-59
TC76538 homologue to GP|6969496|gb|AAF33785.1| cold acclimation ... 151 7e-38
TC78144 similar to PIR|T07078|T07078 cold stress protein SRC1 - ... 99 4e-22
BE248994 similar to GP|16604364|gb| AT5g39570/MIJ24_40 {Arabidop... 52 4e-08
BQ144275 48 1e-06
AA660759 similar to GP|16604364|gb| AT5g39570/MIJ24_40 {Arabidop... 47 2e-06
TC76705 similar to SP|Q07202|CORA_MEDSA Cold and drought-regulat... 43 3e-05
BQ143793 weakly similar to GP|6523547|emb| hydroxyproline-rich g... 42 7e-05
TC76697 40 2e-04
TC92973 weakly similar to GP|12328526|dbj|BAB21184. hypothetical... 40 2e-04
TC91992 similar to GP|8099125|dbj|BAA90497.1 rice EST C27893 cor... 40 2e-04
TC76706 similar to SP|Q07202|CORA_MEDSA Cold and drought-regulat... 40 3e-04
TC76911 similar to GP|13543234|gb|AAH05782.1 Unknown (protein fo... 40 3e-04
TC76707 similar to SP|Q07202|CORA_MEDSA Cold and drought-regulat... 39 4e-04
TC88391 similar to PIR|T03986|T03986 transformer-SR ribonucleopr... 39 5e-04
TC76720 similar to GP|11121502|emb|CAC14888. putative extensin {... 39 6e-04
BQ141909 weakly similar to GP|14253159|emb VMP3 protein {Volvox ... 38 8e-04
TC81816 similar to GP|8096269|dbj|BAA95789.1 KED {Nicotiana taba... 37 0.001
AW690548 similar to SP|Q09134|GRPA Abscisic acid and environment... 31 0.099
>TC76537 homologue to GP|6969496|gb|AAF33785.1| cold acclimation responsive
protein BudCAR5 {Medicago sativa}, partial (87%)
Length = 682
Score = 271 bits (692), Expect = 6e-74
Identities = 121/121 (100%), Positives = 121/121 (100%)
Frame = +3
Query: 1 MAGIMNKIGDALHIGGDKKEGEHKGEQHGHVGGEHHGEYKGEQHGFVGGHGGEHKGEQHG 60
MAGIMNKIGDALHIGGDKKEGEHKGEQHGHVGGEHHGEYKGEQHGFVGGHGGEHKGEQHG
Sbjct: 66 MAGIMNKIGDALHIGGDKKEGEHKGEQHGHVGGEHHGEYKGEQHGFVGGHGGEHKGEQHG 245
Query: 61 FGHGDHKEGHHGEEHKEGFVDKIKDKIHGEGADGEKKKKKEKKKHGEGHEHGHDSSSSDS 120
FGHGDHKEGHHGEEHKEGFVDKIKDKIHGEGADGEKKKKKEKKKHGEGHEHGHDSSSSDS
Sbjct: 246 FGHGDHKEGHHGEEHKEGFVDKIKDKIHGEGADGEKKKKKEKKKHGEGHEHGHDSSSSDS 425
Query: 121 D 121
D
Sbjct: 426 D 428
>TC76542 homologue to GP|6969496|gb|AAF33785.1| cold acclimation responsive
protein BudCAR5 {Medicago sativa}, partial (69%)
Length = 1167
Score = 220 bits (561), Expect = 9e-59
Identities = 98/107 (91%), Positives = 100/107 (92%)
Frame = +2
Query: 1 MAGIMNKIGDALHIGGDKKEGEHKGEQHGHVGGEHHGEYKGEQHGFVGGHGGEHKGEQHG 60
MAGIMNKIGDALHIGGDKKEGEHKGEQHGHVGGEHHGEYKGEQHGFVGGHGGEHKGEQHG
Sbjct: 551 MAGIMNKIGDALHIGGDKKEGEHKGEQHGHVGGEHHGEYKGEQHGFVGGHGGEHKGEQHG 730
Query: 61 FGHGDHKEGHHGEEHKEGFVDKIKDKIHGEGADGEKKKKKEKKKHGE 107
FGHGDHKEGHHGEEHKEGFVDKIKDKIHGEGADGEKK + G+
Sbjct: 731 FGHGDHKEGHHGEEHKEGFVDKIKDKIHGEGADGEKKTSCRIRHEGQ 871
>TC76538 homologue to GP|6969496|gb|AAF33785.1| cold acclimation responsive
protein BudCAR5 {Medicago sativa}, partial (48%)
Length = 499
Score = 151 bits (381), Expect = 7e-38
Identities = 67/67 (100%), Positives = 67/67 (100%)
Frame = +1
Query: 55 KGEQHGFGHGDHKEGHHGEEHKEGFVDKIKDKIHGEGADGEKKKKKEKKKHGEGHEHGHD 114
KGEQHGFGHGDHKEGHHGEEHKEGFVDKIKDKIHGEGADGEKKKKKEKKKHGEGHEHGHD
Sbjct: 49 KGEQHGFGHGDHKEGHHGEEHKEGFVDKIKDKIHGEGADGEKKKKKEKKKHGEGHEHGHD 228
Query: 115 SSSSDSD 121
SSSSDSD
Sbjct: 229 SSSSDSD 249
>TC78144 similar to PIR|T07078|T07078 cold stress protein SRC1 - soybean,
partial (59%)
Length = 698
Score = 99.0 bits (245), Expect = 4e-22
Identities = 65/128 (50%), Positives = 75/128 (57%), Gaps = 7/128 (5%)
Frame = +2
Query: 1 MAGIMNKIGDALHIGGD-KKEGEHKGEQHGHVGGEHHGEYKGEQHGFVGGHGGEHKGEQH 59
MAGI+NKIG+ALH+GG KKE EHKGE+ H +HKGE+
Sbjct: 59 MAGIINKIGEALHVGGGHKKEDEHKGEK---------------------SHDDKHKGEK- 172
Query: 60 GFGHGDHKEGHHGEEHKEGFVDKIKDKIH---GEGAD--GEKKKKKEKKKHGEGHEHGHD 114
H + H GE HKEG VDKIKDKIH GEG + GEKK KK+K K + H H HD
Sbjct: 173 -----SHDDKHKGE-HKEGIVDKIKDKIHGGDGEGHEHKGEKKDKKKKDKKKKEHGHDHD 334
Query: 115 -SSSSDSD 121
SSSSDSD
Sbjct: 335 SSSSSDSD 358
>BE248994 similar to GP|16604364|gb| AT5g39570/MIJ24_40 {Arabidopsis
thaliana}, partial (20%)
Length = 579
Score = 52.4 bits (124), Expect = 4e-08
Identities = 33/116 (28%), Positives = 59/116 (50%), Gaps = 11/116 (9%)
Frame = -2
Query: 15 GGDKKEGEHKGEQHGHVGGEHHGEY------KGEQHGFVGGHGGEHKGEQHGFGHGDHKE 68
GG K+EGE+ G GG GEY K E+ + G+GG + +G G+G +E
Sbjct: 518 GGRKQEGEYGSGYGG--GGAQEGEYGSGYGRKNEESEYGSGYGGRKQESGYGSGYGGAQE 345
Query: 69 GHHGEEHKE-----GFVDKIKDKIHGEGADGEKKKKKEKKKHGEGHEHGHDSSSSD 119
+G +++E G+ + ++ +G G G +K E++ G G ++ ++S S+
Sbjct: 344 SEYGRKNEESGYGSGYGGRKQESGYGSGYGG---RKNEEESEGYGRKNEYESGESE 186
Score = 34.7 bits (78), Expect = 0.009
Identities = 21/100 (21%), Positives = 44/100 (44%), Gaps = 11/100 (11%)
Frame = -2
Query: 32 GGEHHGEYKGEQHGFVGGHGGEHKGEQH--GFGHGDHKEGHHG---------EEHKEGFV 80
G + Y G++ G+GG + ++ G+G G +EG +G E+ G+
Sbjct: 575 GRKQGSGYGGQESEIGSGYGGRKQEGEYGSGYGGGGAQEGEYGSGYGRKNEESEYGSGYG 396
Query: 81 DKIKDKIHGEGADGEKKKKKEKKKHGEGHEHGHDSSSSDS 120
+ ++ +G G G ++ + +K G+ G+ +S
Sbjct: 395 GRKQESGYGSGYGGAQESEYGRKNEESGYGSGYGGRKQES 276
>BQ144275
Length = 790
Score = 47.8 bits (112), Expect = 1e-06
Identities = 30/75 (40%), Positives = 37/75 (49%)
Frame = +2
Query: 1 MAGIMNKIGDALHIGGDKKEGEHKGEQHGHVGGEHHGEYKGEQHGFVGGHGGEHKGEQHG 60
M NK L IGG E+ GE H HV EH + K + H F+ HGG+ G Q G
Sbjct: 59 MVAFFNKTEYPLAIGGYLYICEYVGEGHAHVL*EHICDAKRKAHRFL*RHGGDPCGNQPG 238
Query: 61 FGHGDHKEGHHGEEH 75
G+H +G EEH
Sbjct: 239 TCLGNHFDGLPWEEH 283
>AA660759 similar to GP|16604364|gb| AT5g39570/MIJ24_40 {Arabidopsis
thaliana}, partial (15%)
Length = 570
Score = 47.0 bits (110), Expect = 2e-06
Identities = 35/107 (32%), Positives = 48/107 (44%), Gaps = 2/107 (1%)
Frame = +2
Query: 15 GGDKKEGEHKGEQHGHVGGEHHGEY--KGEQHGFVGGHGGEHKGEQHGFGHGDHKEGHHG 72
GG K+E G G+ GG EY K E+ G+ G+GG + +G G+G G
Sbjct: 26 GGRKQES---GYGSGY-GGAQESEYGRKNEESGYGSGYGGRKQESGYGSGYG----GRKN 181
Query: 73 EEHKEGFVDKIKDKIHGEGADGEKKKKKEKKKHGEGHEHGHDSSSSD 119
EE EG+ K E GE + +KK G G E G+ S+
Sbjct: 182 EEESEGYGRK------NEYESGESEYGSGRKKSGYGEEEGYGXGXSE 304
Score = 36.6 bits (83), Expect = 0.002
Identities = 19/76 (25%), Positives = 41/76 (53%), Gaps = 5/76 (6%)
Frame = +2
Query: 49 GHGGEHKGEQHGFGHGDHKEGHHGEEHKE-----GFVDKIKDKIHGEGADGEKKKKKEKK 103
G+GG + +G G+G +E +G +++E G+ + ++ +G G G +K E++
Sbjct: 20 GYGGRKQESGYGSGYGGAQESEYGRKNEESGYGSGYGGRKQESGYGSGYGG---RKNEEE 190
Query: 104 KHGEGHEHGHDSSSSD 119
G G ++ ++S S+
Sbjct: 191 SEGYGRKNEYESGESE 238
>TC76705 similar to SP|Q07202|CORA_MEDSA Cold and drought-regulated protein
CORA. [Alfalfa] {Medicago sativa}, partial (80%)
Length = 854
Score = 42.7 bits (99), Expect = 3e-05
Identities = 35/119 (29%), Positives = 48/119 (39%), Gaps = 13/119 (10%)
Frame = +2
Query: 6 NKIGDALH----IGGDKKEGEHKGEQHGHVGGEHH-GEYKGEQHGFVGGHGGEHKGEQHG 60
N++ DA + GG G + G + H GG H+ G Y G+ G GG + G HG
Sbjct: 173 NEVNDAKYGGYNHGGYNHGGGYNGGGYNHGGGYHNGGGYHNGGGGYHNGGGGYNGGGGHG 352
Query: 61 FGHGDHKE--GHHGEEHKEGFVDKIKDKIH------GEGADGEKKKKKEKKKHGEGHEH 111
GHG + GH G E + ++K + G G K HG G H
Sbjct: 353 -GHGGYNRGGGHGGRGAAESVAVQTEEKTNEVNDARNGGGGGSFNKGGASYNHGRGSYH 526
Score = 28.5 bits (62), Expect = 0.64
Identities = 18/68 (26%), Positives = 27/68 (39%)
Frame = +2
Query: 10 DALHIGGDKKEGEHKGEQHGHVGGEHHGEYKGEQHGFVGGHGGEHKGEQHGFGHGDHKEG 69
+++ + ++K E ++G GG + HG H G G GHG G
Sbjct: 404 ESVAVQTEEKTNEVNDARNGGGGGSFNKGGASYNHGRGSYHHG*GGYNHGGGGHGGGGHG 583
Query: 70 HHGEEHKE 77
HG E E
Sbjct: 584 SHGAEQTE 607
>BQ143793 weakly similar to GP|6523547|emb| hydroxyproline-rich glycoprotein
DZ-HRGP {Volvox carteri f. nagariensis}, partial (19%)
Length = 1100
Score = 41.6 bits (96), Expect = 7e-05
Identities = 42/132 (31%), Positives = 58/132 (43%), Gaps = 22/132 (16%)
Frame = -1
Query: 3 GIMNKIGDALHIGGDKKEGEHKGEQHGHVGGE------HHGEYKGEQHGFVGGHGGEHKG 56
G N I + + GG G+ K ++ G G G G +G GG GGE G
Sbjct: 395 GAGNGIYEGEYKGGGGGGGKRKKKREGGRGKGKDEII*EKGGGMGRGNGGEGGGGGEGGG 216
Query: 57 EQHGFGHGDHKEGHHGEEHKEGFVDKI---KDKIHGEG------------ADGEKKKKKE 101
+ G G GD E+ K+G DKI K + GEG +G KK+++E
Sbjct: 215 REWGGGGGDK*SLKRKEKEKDG-GDKIG*GKGRGGGEGGMDVMGNGGGRRGEGRKKEERE 39
Query: 102 KK-KHGEGHEHG 112
K+ + GEG E G
Sbjct: 38 KRGEEGEGKEGG 3
Score = 39.7 bits (91), Expect = 3e-04
Identities = 28/85 (32%), Positives = 41/85 (47%)
Frame = -2
Query: 18 KKEGEHKGEQHGHVGGEHHGEYKGEQHGFVGGHGGEHKGEQHGFGHGDHKEGHHGEEHKE 77
K++GE K EQ GG+ + +GE G G G +G++ G K+G +E
Sbjct: 544 KRKGESKWEQGQGEGGDKRKKGRGEGKG---GKGKTQRGKRGG------KKGGRRRGRRE 392
Query: 78 GFVDKIKDKIHGEGADGEKKKKKEK 102
+ +K I G GEK KKKE+
Sbjct: 391 REMGYMKGSIRGGEGGGEKGKKKER 317
Score = 35.4 bits (80), Expect = 0.005
Identities = 28/92 (30%), Positives = 39/92 (41%)
Frame = -3
Query: 17 DKKEGEHKGEQHGHVGGEHHGEYKGEQHGFVGGHGGEHKGEQHGFGHGDHKEGHHGEEHK 76
D E G + G V G G G E +G++ G G ++G GEE
Sbjct: 570 DGANEERWGRERGRVNGNRGKGRGGTSERKEEGKEKEERGKRKE-GKGGERKGEEGEEEG 394
Query: 77 EGFVDKIKDKIHGEGADGEKKKKKEKKKHGEG 108
G D I + G G G KK+KK++ G+G
Sbjct: 393 SGKWD-I*RGV*GGGRGGGKKEKKKRGWEGKG 301
>TC76697
Length = 922
Score = 40.4 bits (93), Expect = 2e-04
Identities = 42/152 (27%), Positives = 56/152 (36%), Gaps = 50/152 (32%)
Frame = +1
Query: 11 ALHI--GGDKKEGEHK-GEQHGH---VGGEH---HGEYKGEQHGFVG---GHGG------ 52
A+H+ GG KE +H + HGH +GG H H + HGF GHG
Sbjct: 205 AMHMKPGGFGKESDHSFSKHHGHGSNIGGNHNMFHQDSMSNGHGFGNNNHGHGNGQKFPF 384
Query: 53 --------EHKGEQHGFGHGDHKEGHHG--EEHKEGFVDKIKDKIHGEGADGEKKKKKEK 102
H G F H H GH+ EH+E + K++ G GA + + E+
Sbjct: 385 GATNKHSPHHGGGVRPFNH--HGGGHNDYVSEHEEYEFEAYKEECVGSGASKMDEMRYER 558
Query: 103 ----------------------KKHGEGHEHG 112
K HG GH HG
Sbjct: 559 HGTYGGDVHYANPYGYNNNNRIKPHGHGHGHG 654
Score = 32.0 bits (71), Expect = 0.058
Identities = 19/54 (35%), Positives = 24/54 (44%), Gaps = 2/54 (3%)
Frame = +1
Query: 16 GDKKEGEHKGEQHGHVGGEHH--GEYKGEQHGFVGGHGGEHKGEQHGFGHGDHK 67
G K E + E+HG GG+ H Y + + HG HG GHG HK
Sbjct: 523 GASKMDEMRYERHGTYGGDVHYANPYGYNNNNRIKPHG-------HGHGHGSHK 663
>TC92973 weakly similar to GP|12328526|dbj|BAB21184. hypothetical
protein~similar to Arabidopsis thaliana chromosome 3
F28L1.7, partial (15%)
Length = 683
Score = 40.4 bits (93), Expect = 2e-04
Identities = 31/95 (32%), Positives = 43/95 (44%), Gaps = 3/95 (3%)
Frame = +2
Query: 17 DKKEGEHKGEQHGHVGGEHHGEYKGE-QHGFVGGHGG--EHKGEQHGFGHGDHKEGHHGE 73
DKK + KGE + E + K +H H G + G+ G DHKEG++G
Sbjct: 347 DKKSSKSKGETENIINKEIKEDQKNSTEHIGESSHEGCIDDAGDD-GEEDSDHKEGNNG- 520
Query: 74 EHKEGFVDKIKDKIHGEGADGEKKKKKEKKKHGEG 108
DK + + G KKKKK+KKK+ G
Sbjct: 521 -----------DKNNNKSEGGAKKKKKKKKKNXSG 592
>TC91992 similar to GP|8099125|dbj|BAA90497.1 rice EST C27893 corresponds to
a region of the predicated gene; unknown protein {Oryza
sativa}, partial (28%)
Length = 733
Score = 40.0 bits (92), Expect = 2e-04
Identities = 16/35 (45%), Positives = 20/35 (56%)
Frame = +1
Query: 44 HGFVGGHGGEHKGEQHGFGHGDHKEGHHGEEHKEG 78
H GHGG H G +G+GHG +K G G+ K G
Sbjct: 391 HHLSHGHGGYHHGYGYGYGHGKYKHGKFGKRWKHG 495
Score = 33.5 bits (75), Expect = 0.020
Identities = 24/69 (34%), Positives = 28/69 (39%), Gaps = 1/69 (1%)
Frame = +1
Query: 9 GDALHIGGDKKEGEHKGEQHGHVGGEHHGEYKGEQHGFVGGHGGEHKGEQHG-FGHGDHK 67
G A G H G HG+ G HG+YK HG K +HG FG G K
Sbjct: 367 GAAAAYGAHHLSHGHGGYHHGYGYGYGHGKYK---------HGKFGKRWKHGRFGFGKFK 519
Query: 68 EGHHGEEHK 76
G G+ K
Sbjct: 520 HGKFGKRWK 546
>TC76706 similar to SP|Q07202|CORA_MEDSA Cold and drought-regulated protein
CORA. [Alfalfa] {Medicago sativa}, partial (94%)
Length = 1034
Score = 39.7 bits (91), Expect = 3e-04
Identities = 26/83 (31%), Positives = 33/83 (39%), Gaps = 3/83 (3%)
Frame = +1
Query: 6 NKIGDALHIGGDKKEG---EHKGEQHGHVGGEHHGEYKGEQHGFVGGHGGEHKGEQHGFG 62
N++ DA + GG G H G + H GG +H G HG G G
Sbjct: 457 NEVNDAKYGGGSYNHGGSYNHGGGSYNHGGGSYHHGGGGYNHG--------------GGG 594
Query: 63 HGDHKEGHHGEEHKEGFVDKIKD 85
HG H G HG E DK ++
Sbjct: 595 HGGHGGGGHGGHGAEQTEDKTQN 663
Score = 39.3 bits (90), Expect = 4e-04
Identities = 35/127 (27%), Positives = 50/127 (38%), Gaps = 17/127 (13%)
Frame = +1
Query: 6 NKIGDALH----IGGDKKEGEHKGEQHGHVGGEHHG-----------EYKGEQHGFVGGH 50
N++ DA + GG G + G + H GG H+G + G + GGH
Sbjct: 163 NEVNDAKYGGYNHGGYNHGGGYNGGGYNHGGGYHNGGGGYHNGGGGYNHGGGGYNGGGGH 342
Query: 51 GGEHKGEQHGFGHGDHK--EGHHGEEHKEGFVDKIKDKIHGEGADGEKKKKKEKKKHGEG 108
GG H G G GHG + GH G E + ++K + + K HG
Sbjct: 343 GG-HGGYNGGGGHGGYNGGGGHGGHGAAESVAVQTEEKTN---EVNDAKYGGGSYNHGGS 510
Query: 109 HEHGHDS 115
+ HG S
Sbjct: 511 YNHGGGS 531
Score = 26.9 bits (58), Expect = 1.9
Identities = 12/37 (32%), Positives = 14/37 (37%)
Frame = -2
Query: 23 HKGEQHGHVGGEHHGEYKGEQHGFVGGHGGEHKGEQH 59
H G H H G HH G+ H H+G H
Sbjct: 313 HHGCNHLHRCGNHHHRCGNHHRGYNLHHCNHHRGYNH 203
>TC76911 similar to GP|13543234|gb|AAH05782.1 Unknown (protein for
MGC:12025) {Mus musculus}, partial (57%)
Length = 1065
Score = 39.7 bits (91), Expect = 3e-04
Identities = 21/48 (43%), Positives = 27/48 (55%), Gaps = 1/48 (2%)
Frame = +2
Query: 15 GGDKKEGEHKGEQHGHVGGEHHGEYKGEQHG-FVGGHGGEHKGEQHGF 61
GG G H G GH + HG++KG +HG F G G+H G +HGF
Sbjct: 536 GGYAHGGSHMGHM-GHGKFKQHGKFKGGKHGKFKHGKFGKHGGGKHGF 676
>TC76707 similar to SP|Q07202|CORA_MEDSA Cold and drought-regulated protein
CORA. [Alfalfa] {Medicago sativa}, partial (87%)
Length = 780
Score = 39.3 bits (90), Expect = 4e-04
Identities = 26/72 (36%), Positives = 31/72 (42%)
Frame = +1
Query: 6 NKIGDALHIGGDKKEGEHKGEQHGHVGGEHHGEYKGEQHGFVGGHGGEHKGEQHGFGHGD 65
N++ DA + GG H G + H GG +H GG G H G HG GHG
Sbjct: 508 NEVNDAKYGGGYN----HGGGSYNHGGGSYHH----------GGGGYNHGGGGHG-GHGG 642
Query: 66 HKEGHHGEEHKE 77
G HG E E
Sbjct: 643 GGHGGHGAEQTE 678
Score = 34.3 bits (77), Expect = 0.012
Identities = 32/118 (27%), Positives = 42/118 (35%), Gaps = 15/118 (12%)
Frame = +1
Query: 13 HIGGDKKEGEHKGEQHGHVGGEHHGEYKGEQHGFVGGHGG--EHKGEQHGFGHGDHKEGH 70
H GG G G + H GG ++ G HG G +GG H G + G G + G
Sbjct: 217 HGGGGYNNG---GGGYNHGGGGYNNGGGGYNHGGGGYNGGGYNHGGGGYNHGGGGYNHGG 387
Query: 71 HGEEHKEGFVDKIKDKIHGEGADG-------------EKKKKKEKKKHGEGHEHGHDS 115
G G +G G G EK + K+G G+ HG S
Sbjct: 388 GGYNGGGGHGGHGGGGYNGGGGHGGHGAAESVAVQTEEKTNEVNDAKYGGGYNHGGGS 561
>TC88391 similar to PIR|T03986|T03986 transformer-SR ribonucleoprotein -
common tobacco (fragment), partial (42%)
Length = 1037
Score = 38.9 bits (89), Expect = 5e-04
Identities = 13/41 (31%), Positives = 26/41 (62%), Gaps = 1/41 (2%)
Frame = +2
Query: 21 GEHKGEQHGHVGGEHHGEYKGEQHG-FVGGHGGEHKGEQHG 60
G+H+G+ G G+H G+++G+ G + H G+++G+ G
Sbjct: 578 GDHRGDHRGDFRGDHRGDFRGDHRGDYRSDHRGDYRGDYRG 700
Score = 33.9 bits (76), Expect = 0.015
Identities = 21/92 (22%), Positives = 33/92 (35%)
Frame = +2
Query: 29 GHVGGEHHGEYKGEQHGFVGGHGGEHKGEQHGFGHGDHKEGHHGEEHKEGFVDKIKDKIH 88
GH G + G + G H G+ +G+ G GDH+ + + + D D+
Sbjct: 533 GHYLGLKNTRDYGPRGDHRGDHRGDFRGDHRGDFRGDHRGDYRSDHRGDYRGDYRGDRGR 712
Query: 89 GEGADGEKKKKKEKKKHGEGHEHGHDSSSSDS 120
G G + + GHD S S
Sbjct: 713 NRGGSGRVDYSDRRSPRRSPYRGGHDHSPQRS 808
Score = 33.1 bits (74), Expect = 0.026
Identities = 15/54 (27%), Positives = 29/54 (52%), Gaps = 1/54 (1%)
Frame = +2
Query: 16 GDKKEGEHKGEQHGHVGGEHHGEYKGEQHG-FVGGHGGEHKGEQHGFGHGDHKE 68
GD + G+ +G+ G G+H G+Y+ + G + G + G+ + G G D+ +
Sbjct: 590 GDHR-GDFRGDHRGDFRGDHRGDYRSDHRGDYRGDYRGDRGRNRGGSGRVDYSD 748
Score = 32.0 bits (71), Expect = 0.058
Identities = 13/62 (20%), Positives = 29/62 (45%)
Frame = +2
Query: 21 GEHKGEQHGHVGGEHHGEYKGEQHGFVGGHGGEHKGEQHGFGHGDHKEGHHGEEHKEGFV 80
G+H+G+ G++ G+Y+G++ GG G ++ ++ GH + +V
Sbjct: 638 GDHRGDYRSDHRGDYRGDYRGDRGRNRGGSGRVDYSDRRSPRRSPYRGGHDHSPQRSPYV 817
Query: 81 DK 82
+
Sbjct: 818 GR 823
Score = 30.4 bits (67), Expect = 0.17
Identities = 14/47 (29%), Positives = 18/47 (37%)
Frame = -1
Query: 35 HHGEYKGEQHGFVGGHGGEHKGEQHGFGHGDHKEGHHGEEHKEGFVD 81
HH + H +H H H H +GHH E H +G D
Sbjct: 722 HHDSFPDHLGSHRDSHHDDHCDSHHDDHHESHHDGHH-ESHHDGHRD 585
Score = 26.6 bits (57), Expect = 2.4
Identities = 16/51 (31%), Positives = 21/51 (40%)
Frame = -1
Query: 21 GEHKGEQHGHVGGEHHGEYKGEQHGFVGGHGGEHKGEQHGFGHGDHKEGHH 71
G H+ H HH ++ E H H G H E H H H++ HH
Sbjct: 695 GSHRDSHHDDHCDSHHDDHH-ESH-----HDGHH--ESH---HDGHRDDHH 576
Score = 25.4 bits (54), Expect = 5.5
Identities = 18/74 (24%), Positives = 25/74 (33%)
Frame = -1
Query: 36 HGEYKGEQHGFVGGHGGEHKGEQHGFGHGDHKEGHHGEEHKEGFVDKIKDKIHGEGADGE 95
+ ++ H H G H+ H H + HH E H +G H DG
Sbjct: 743 NNQHDQNHHDSFPDHLGSHRDSHHDDHCDSHHDDHH-ESHHDGH--------HESHHDG- 594
Query: 96 KKKKKEKKKHGEGH 109
+ H EGH
Sbjct: 593 -----HRDDHHEGH 567
>TC76720 similar to GP|11121502|emb|CAC14888. putative extensin {Nicotiana
sylvestris}, partial (51%)
Length = 1290
Score = 38.5 bits (88), Expect = 6e-04
Identities = 31/92 (33%), Positives = 40/92 (42%), Gaps = 2/92 (2%)
Frame = -2
Query: 15 GGDKKEGEHKGEQHGHVGGEHHGEYK--GEQHGFVGGHGGEHKGEQHGFGHGDHKEGHHG 72
G EGE +GE+ G + GE G G+ G G GGE GE G G G EG
Sbjct: 761 GAGDDEGEGEGEEEGELSGEGAGGESE*GDGVGVGVGAGGEFAGEGVGIGVG---EGEFS 591
Query: 73 EEHKEGFVDKIKDKIHGEGADGEKKKKKEKKK 104
+E G GE A+ KKE+++
Sbjct: 590 DEFVAGVG-------AGESAEEAMLTKKERRR 516
>BQ141909 weakly similar to GP|14253159|emb VMP3 protein {Volvox carteri f.
nagariensis}, partial (13%)
Length = 1223
Score = 38.1 bits (87), Expect = 8e-04
Identities = 31/88 (35%), Positives = 42/88 (47%)
Frame = +3
Query: 25 GEQHGHVGGEHHGEYKGEQHGFVGGHGGEHKGEQHGFGHGDHKEGHHGEEHKEGFVDKIK 84
G + G GGE G +GE+ G GG GGE G G G ++G GEE KEG
Sbjct: 186 GGRGGRGGGERRGGRRGEEGG-GGGGGGE------GGGGGGGRKG--GEEQKEG------ 320
Query: 85 DKIHGEGADGEKKKKKEKKKHGEGHEHG 112
G +GE+++ ++K EG G
Sbjct: 321 ----GSVREGEERRGAAREK*REGMGQG 392
>TC81816 similar to GP|8096269|dbj|BAA95789.1 KED {Nicotiana tabacum},
partial (13%)
Length = 663
Score = 37.4 bits (85), Expect = 0.001
Identities = 34/106 (32%), Positives = 48/106 (45%), Gaps = 8/106 (7%)
Frame = +3
Query: 15 GGDKKEGEHKGEQHGHV---GGEHHGEYKGEQHGFVGGHGGEHKGEQHGFGHGDH-KEGH 70
G DKK+ E K ++ G E G+ K ++ + K E+ G D K+G
Sbjct: 234 GKDKKDKEKKKKEKKEENVKGEEEDGDEKKDKE--------KKKKEKKEKGKEDKDKDGE 389
Query: 71 HGEEHKEGFVDKIKDKIHGEGADGEKKK----KKEKKKHGEGHEHG 112
+ K+ K K++ EG DG KKK KKEKKK + E G
Sbjct: 390 EKKSKKDKEKKKDKNEDDDEGEDGSKKKKNKDKKEKKKEEDEKEEG 527
Score = 35.4 bits (80), Expect = 0.005
Identities = 19/57 (33%), Positives = 31/57 (54%)
Frame = +3
Query: 65 DHKEGHHGEEHKEGFVDKIKDKIHGEGADGEKKKKKEKKKHGEGHEHGHDSSSSDSD 121
D EG ++ ++ +K ++ + GE DG++KK KEKKK E E G + D +
Sbjct: 222 DVDEGKDKKDKEKKKKEKKEENVKGEEEDGDEKKDKEKKKK-EKKEKGKEDKDKDGE 389
>AW690548 similar to SP|Q09134|GRPA Abscisic acid and environmental stress
inducible protein. [Sickle medic] {Medicago falcata},
partial (68%)
Length = 599
Score = 31.2 bits (69), Expect = 0.099
Identities = 29/108 (26%), Positives = 34/108 (30%), Gaps = 41/108 (37%)
Frame = +1
Query: 6 NKIGDALHIGGDKKEGEHKGEQHGHVGGEHHGEYKGEQHGFVGGH--------------- 50
N++ DA + GG H G H + GG HHG G HG G H
Sbjct: 151 NEVNDAKYGGGGNY---HNGGGHYYGGGSHHGG-GGSHHGGGGCHYYCHGHCCSYAEFVA 318
Query: 51 ------------------------GGEHKGEQHGFGHGDHKEG--HHG 72
G H G H +G G H G HHG
Sbjct: 319 VQTEEKTNEVNDAKYGGGGYHNGGGNYHNGGGHYYGGGSHGGGGSHHG 462
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.307 0.137 0.417
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,603,068
Number of Sequences: 36976
Number of extensions: 81332
Number of successful extensions: 887
Number of sequences better than 10.0: 190
Number of HSP's better than 10.0 without gapping: 637
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 753
length of query: 121
length of database: 9,014,727
effective HSP length: 84
effective length of query: 37
effective length of database: 5,908,743
effective search space: 218623491
effective search space used: 218623491
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 43 (21.9 bits)
S2: 52 (24.6 bits)
Medicago: description of AC137822.11


